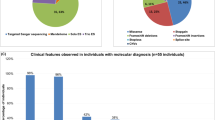
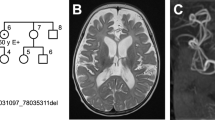
Abstract
Intellectual disability (ID) comprises a large group of heterogeneous disorders, often without a known molecular cause. X-linked ID accounts for 5–10% of male ID cases. We investigated a large, three-generation family with mild ID and behavior problems in five males and one female, with a segregation suggestive for X-linked inheritance. Linkage analysis mapped a disease locus to a 7.6 Mb candidate region on the X-chromosome (LOD score 3.3). Whole-genome sequencing identified a 2 bp insertion in exon 2 of the chromosome X open reading frame 56 gene (CXorf56), resulting in a premature stop codon. This insertion was present in all intellectually impaired individuals and carrier females. Additionally, X-inactivation status showed skewed methylation patterns favoring the inactivation of the mutated allele in the unaffected carrier females. We demonstrate that the insertion leads to nonsense-mediated decay and that CXorf56 mRNA expression is reduced in the impaired males and female. In murine brain slices and primary hippocampal neuronal cultures, CXorf56 protein was present and localized in the nucleus, cell soma, dendrites, and dendritic spines. Although no other families have been identified with pathogenic variants in CXorf56, these results suggest that CXorf56 is the causative gene in this family, and thus a novel candidate gene for X-linked ID with behavior problems.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Vissers LE, Gilissen C, Veltman JA. Genetic studies in intellectual disability and related disorders. Nat Rev Genet. 2016;17:9–18.
Willemsen MH, Kleefstra T. Making headway with genetic diagnostics of intellectual disabilities. Clin Genet. 2014;85:101–10.
Gecz J, Shoubridge C, Corbett M. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 2009;25:308–16.
Lubs HA, Stevenson RE, Schwartz CE. Fragile X and X-linked intellectual disability: four decades of discovery. Am J Hum Genet. 2012;90:579–90.
Ropers HH. Genetics of early onset cognitive impairment. Annu Rev Genom Hum Genet. 2010;11:161–87.
Verkerk AJ, Pieretti M, Sutcliffe JS, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–14.
Kaufman L, Ayub M, Vincent JB. The genetic basis of non-syndromic intellectual disability: a review. J Neurodev Disord. 2010;2:182–209.
De Vries BB, Winter R, Schinzel A, van Ravenswaaij-Arts C. Telomeres: a diagnosis at the end of the chromosomes. J Med Genet. 2003;40:385–98.
Rooms L, Vandeweyer G, Reyniers E, et al. Array-based MLPA to detect recurrent copy number variations in patients with idiopathic mental retardation. Am J Med Genet A. 2011;155a:343–8.
Vissers LE, de Vries BB, Veltman JA. Genomic microarrays in mental retardation: from copy number variation to gene, from research to diagnosis. J Med Genet. 2010;47:289–97.
Whibley AC, Plagnol V, Tarpey PS, et al. Fine-scale survey of X chromosome copy number variants and indels underlying intellectual disability. Am J Hum Genet. 2010;87:173–88.
Verkerk AJ, Schot R, Dumee B, et al. Mutation in the AP4M1 gene provides a model for neuroaxonal injury in cerebral palsy. Am J Hum Genet. 2009;85:40–52.
Musante L, Ropers HH. Genetics of recessive cognitive disorders. Trends Genet. 2014;30:32–9.
Rabbani B, Mahdieh N, Hosomichi K, Nakaoka H, Inoue I. Next-generation sequencing: impact of exome sequencing in characterizing Mendelian disorders. J Hum Genet. 2012;57:621–32.
Tarpey PS, Smith R, Pleasance E, et al. A systematic, large-scale resequencing screen of X-chromosome coding exons in mental retardation. Nat Genet. 2009;41:535–43.
Vissers LE, de Ligt J, Gilissen C, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12.
Kuhlenbaumer G, Hullmann J, Appenzeller S. Novel genomic techniques open new avenues in the analysis of monogenic disorders. Hum Mutat. 2011;32:144–51.
Hu H, Haas SA, Chelly J, et al. X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry. 2016;21:133–48.
Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat Genet. 2000;25:12–3.
Lindner TH, Hoffmann K. easyLINKAGE: a PERL script for easy and automated two-/multi-point linkage analyses. Bioinformatics. 2005;21:405–7.
Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81.
Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet. 1992;51:1229–39.
de Brouwer AP, van Bokhoven H, Kremer H. Comparison of 12 reference genes for normalization of gene expression levels in Epstein-Barr virus-transformed lymphoblastoid cell lines and fibroblasts. Mol Diagn Ther. 2006;10:197–204.
Popp MW, Maquat LE. Leveraging rules of nonsense-mediated mRNA decay for genome engineering and personalized medicine. Cell. 2016;165:1319–22.
Schweingruber C, Rufener SC, Zund D, Yamashita A, Muhlemann O. Nonsense-mediated mRNA decay—mechanisms of substrate mRNA recognition and degradation in mammalian cells. Biochim Biophys Acta. 2013;1829:612–23.
Seelaar H, Schelhaas HJ, Azmani A, et al. TDP-43 pathology in familial frontotemporal dementia and motor neuron disease without Progranulin mutations. Brain. 2007;130:1375–85.
de Vrij FM, Levenga J, van der Linde HC, et al. Rescue of behavioral phenotype and neuronal protrusion morphology in Fmr1 KO mice. Neurobiol Dis. 2008;31:127–32.
Raven J. The Raven’s progressive matrices: change and stability over culture and time. Cogn Psychol. 2000;41:1–48.
Nelson JM, Canivez GL, Watkins MW. Structural and incremental validity of the Wechsler Adult Intelligence Scale-Fourth Edition with a clinical sample. Psychol Assess. 2013;25:618–30.
Kaufman AS, Horn JL. Age changes on tests of fluid and crystallized ability for women and men on the Kaufman Adolescent and Adult Intelligence Test (KAIT) at ages 17-94 years. Arch Clin Neuropsychol. 1996;11:97–121.
Collins JS, Schwartz CE. Detecting polymorphisms and mutations in candidate genes. Am J Hum Genet. 2002;71:1251–2.
Noensie EN, Dietz HC. A strategy for disease gene identification through nonsense-mediated mRNA decay inhibition. Nat Biotechnol. 2001;19:434–9.
Fieremans N, Van Esch H, Holvoet M, et al. Identification of intellectual disability genes in female patients with a skewed X-inactivation pattern. Hum Mutat. 2016;37:804–11.
Orstavik KH, Orstavik RE, Naumova AK, et al. X chromosome inactivation in carriers of Barth syndrome. Am J Hum Genet. 1998;63:1457–63.
Willard HF. X chromosome inactivation and X-linked mental retardation. Am J Med Genet. 1996;64:21–6.
de Vries BB, Wiegers AM, Smits AP, et al. Mental status of females with an FMR1 gene full mutation. Am J Hum Genet. 1996;58:1025–32.
de Brouwer AP, Yntema HG, Kleefstra T, et al. Mutation frequencies of X-linked mental retardation genes in families from the EuroMRX consortium. Hum Mutat. 2007;28:207–8.
Tzschach A, Grasshoff U, Beck-Woedl S, et al. Next-generation sequencing in X-linked intellectual disability. Eur J Hum Genet. 2015;23:1513–8.
Tahirovic S, Bradke F. Neuronal polarity. Cold Spring Harb Perspect Biol. 2009;1:a001644.
Ewing RM, Chu P, Elisma F, et al. Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol Syst Biol. 2007;3:89.
An J, Nakajima T, Shibata H, Arimura T, Yasunami M, Kimura A. A novel link of HLA locus to the regulation of immunity and infection: NFKBIL1 regulates alternative splicing of human immune-related genes and influenza virus M gene. J Autoimmun. 2013;47:25–33.
Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–6.
Cox BJ, Vollmer M, Tamplin O, et al. Phenotypic annotation of the mouse X chromosome. Genome Res. 2010;20:1154–64.
Agafonov DE, Deckert J, Wolf E, et al. Semiquantitative proteomic analysis of the human spliceosome via a novel two-dimensional gel electrophoresis method. Mol Cell Biol. 2011;31:2667–82.
Najmabadi H, Hu H, Garshasbi M, et al. Deep sequencing reveals 50 novel genes for recessive cognitive disorders. Nature. 2011;478:57–63.
Nascimento RM, Otto PA, de Brouwer AP, Vianna-Morgante AM. UBE2A, which encodes a ubiquitin-conjugating enzyme, is mutated in a novel X-linked mental retardation syndrome. Am J Hum Genet. 2006;79:549–55.
Ramakers GJ. Rho proteins, mental retardation and the cellular basis of cognition. Trends Neurosci. 2002;25:191–9.
Thunstrom S, Sodermark L, Ivarsson L, Samuelsson L, Stefanova M. UBE2A deficiency syndrome: a report of two unrelated cases with large Xq24 deletions encompassing UBE2A gene. Am J Med Genet A. 2015;167a:204–10.
Cooper GM, Coe BP, Girirajan S, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46.
Acknowledgements
We thank the family for their continuous cooperation, their endless patience, perseverance, and trust. We thank Diana van Rongen-Nispen and Lorette Dubbel-Hulsman for cell culture, Dicky Halley and Hans van Bokhoven for availability of DNA samples, and Edith Friesema and Theo Visser for supporting the project. Tom de Vries Lentsch is acknowledged for excellent graphical support. We thank Willem Verhoeven and collaborators of the Vincent van Gogh institute for performing IQ tests of family members.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Verkerk, A.J.M.H., Zeidler, S., Breedveld, G. et al. CXorf56, a dendritic neuronal protein, identified as a new candidate gene for X-linked intellectual disability. Eur J Hum Genet 26, 552–560 (2018). https://doi.org/10.1038/s41431-017-0051-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-017-0051-9
This article is cited by
-
Inhibition of CXorf56 promotes PARP inhibitor-induced cytotoxicity in triple-negative breast cancer
npj Breast Cancer (2023)
-
Emerging roles of spliceosome in cancer and immunity
Protein & Cell (2022)
-
Novel clinical and genetic insight into CXorf56-associated intellectual disability
European Journal of Human Genetics (2020)