Abstract
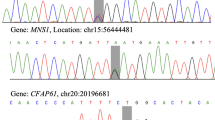
In affected members of a consanguineous family, a syndrome, which is concurrence of set of medical signs, is often observed and commonly assumed to have arisen from pleiotropy, i.e., the phenomenon of a single gene variant affecting multiple traits. We detected six sibs afflicted with a unique combination of digit malformation that includes brachydactyly, symphalangism and zygodactyly plus infertility in males owing to azoospermia, sperm immotility or necrospermia, which we hypothesised to have arisen from a defect in a single gene. We mapped the disease locus and by exome sequencing identified in patients homozygous missense variants bone morphogenetic protein receptor type IB (BMPR1B) c.640C>T (p.(Arg214Cys)) and alpha-2 pyruvate dehydrogenase (PDHA2) c.679A>G (p.(Met227Val)). Structural protein modelling, protein sequence conservation and in silico analysis indicate that both variants affect protein function. BMPR1B is known to be responsible for autosomal dominant brachydactyly and autosomal recessive acromesomelic chondrodysplasia. Our findings show that also recessive complex digit malformation can be caused by BMPR1B variant and not all biallelic BMPR1B variants cause acromesomelic dysplasia. PDHA2 is a novel candidate gene for male infertility; the protein product is a mitochondrial enzyme with highest expression in ejaculated sperm. Our findings are a unique example of two linked variants, ~ 711 Kb apart, in different genes that together manifest as a novel syndrome. They demonstrate that exome sequencing and not candidate gene approach should be employed in disease gene hunt, defining new diseases and genetic testing, to rule out the coincidental presence of two variants contributing together to the phenotype, which may be discerned as a novel disease.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Demirhan O, Turkmen S, Schwabe GC, et al. A homozygous BMPR1B mutation causes a new subtype of acromesomelic chondrodysplasia with genital anomalies. J Med Genet. 2005;42:314–7.
Graul-Neumann LM, Deichsel A, Wille U, et al. Homozygous missense and nonsense mutations in BMPR1B cause acromesomelic chondrodysplasia-type Grebe. Eur J Hum Genet. 2014;22:726–33.
Stange K, Désir J, Kakar N, et al. A hypomorphic BMPR1B mutation causes du Pan acromesomelic dysplasia. Orphanet J Rare Dis. 2015;10:1.
Chalmel F, Rolland AD. Linking transcriptomics and proteomics in spermatogenesis. Reproduction. 2015;150:R149–R157.
Dahl H-H, Brown R, Hutchison W, Maragos C, Brown G. A testis-specific form of the human pyruvate dehydrogenase E1α subunit is coded for by an intronless gene on chromosome 4. Genomics. 1990;8:225–32.
Pinheiro A, Silva MJ, Graca I, et al. Pyruvate dehydrogenase complex: mRNA and protein expression patterns of E1 alpha subunit genes in human spermatogenesis. Gene. 2012;506:173–8.
Ficarro S, Chertihin O, Westbrook VA, et al. Phosphoproteome analysis of capacitated human sperm evidence of tyrosine phosphorylation of a kinase-anchoring protein 3 and valosin-containing protein/p97 during capacitation. J Biol Chem. 2003;278:11579–89.
Chang X, Wang K. wANNOVAR: annotating genetic variants for personal genomes via the web. J Med Genet. 2012;49:433–6.
Chun S, Fay JC. Identification of deleterious mutations within three human genomes. Genome Res. 2009;19:1553–61.
Dong C, Wei P, Jian X, et al. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24:2125–37.
Shihab HA, Gough J, Cooper DN, Day IN, Gaunt TR. Predicting the functional consequences of cancer-associated amino acid substitutions. Bioinformatics. 2013;29:1504–10. btt182
Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–W457.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2.
Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Ayhan Ö, Balkan M, Guven A, et al. Truncating mutations in TAF4B and ZMYND15 causing recessive azoospermia. J Med Genet. 2014;51:239–44.
Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–14.
Kearse M, Moir R, Wilson A, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9.
Simossis VA, Heringa J. PRALINE: a multiple sequence alignment toolbox that integrates homology-extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294.
Kato M, Wynn RM, Chuang JL, et al. Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops. Structure. 2008;16:1849–59.
Korotchkina LG, Sidhu S, Patel MS. Characterization of testis-specific isoenzyme of human pyruvate dehydrogenase. J Biol Chem. 2006;281:9688–96.
Muller YA, Lindqvist Y, Furey W, Schulz GE, Jordan F, Schneider G. A thiamin diphosphate binding fold revealed by comparison of the crystal structures of transketolase, pyruvate oxidase and pyruvate decarboxylase. Structure. 1993;1:95–103.
Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–49.
Kumar V, Rangaraj N, Shivaji S. Activity of pyruvate dehydrogenase A (PDHA) in hamster spermatozoa correlates positively with hyperactivation and is associated with sperm capacitation. Biol Reprod. 2006;75:767–77.
Kolobova E, Tuganova A, Boulatnikov I, Popov KM. Regulation of pyruvate dehydrogenase activity through phosphorylation at multiple sites. Biochem J. 2001;358:69–77.
Badura-Stronka M, Mroz D, Beighton P, et al. Novel mutation in the BMPR1B gene (R486L) in a polish family and further delineation of the phenotypic features of BMPR1B-related brachydactyly. Birth Defects Res Part A-Clin Mol Teratol. 2015;103:567–72.
Lehmann K, Seemann P, Stricker S, et al. Mutations in bone morphogenetic protein receptor 1B cause brachydactyly type A2. Proc Natl Acad Sci USA. 2003;100:12277–82.
Racacho L, Byrnes AM, MacDonald H, et al. Two novel disease-causing variants in BMPR1B are associated with brachydactyly type A1. Eur J Hum Genet. 2015;23:1640–5.
Wilton LJ, Temple-Smith PD, Baker HW, de Kretser DM. Human male infertility caused by degeneration and death of sperm in the epididymis. Fertil Steril. 1988;49:1052–8.
Holyoake AJ, McHugh P, Wu M, et al. High incidence of single nucleotide substitutions in the mitochondrial genome is associated with poor semen parameters in men. Int J Androl. 2001;24:175–82.
Kao SH, Chao HT, Liu HW, Liao TL, Wei YH. Sperm mitochondrial DNA depletion in men with asthenospermia. Fertil Steril. 2004;82:66–73.
St John JC, Jokhi RP, Barratt CLR. The impact of mitochondrial genetics on male infertility. Int J Androl. 2005;28:65–73.
Thangaraj K, Joshi MB, Reddy AG, Rasalkar AA, Singh L. Sperm mitochondrial mutations as a cause of low sperm motility. J Androl. 2003;24:388–92.
Avenarius MR, Hildebrand MS, Zhang Y, et al. Human male infertility caused by mutations in the CATSPER1 channel protein. Am J Hum Genet. 2009;84:505–10.
Maor-Sagie E, Cinnamon Y, Yaacov B, et al. Deleterious mutation in SYCE1 is associated with non-obstructive azoospermia. J Assist Reprod Genet. 2015;32:887–91.
Acknowledgements
We thank the family members for their cooperation. This study was supported by Scientific and Technologic Research Council of Turkey (TÜBİTAK) grant 114Z829 and Boğaziçi University Research Fund grant 10860. UW is supported by FONDECYT no. 1150743.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Compliance with ethical standards
The study protocol was approved by the Ethical Review Committee of Quaid-i-Azam University and the Boğaziçi University Institutional Review Board for Research with Human Participants.
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
41431_2018_121_MOESM1_ESM.docx
Supplementary Material: Supplementary Table 1, Supplementary Figure 1, Supplementary Figure 2, Supplementary Figure 3 and Supplementary Figure 4
Rights and permissions
About this article
Cite this article
Yıldırım, Y., Ouriachi, T., Woehlbier, U. et al. Linked homozygous BMPR1B and PDHA2 variants in a consanguineous family with complex digit malformation and male infertility. Eur J Hum Genet 26, 876–885 (2018). https://doi.org/10.1038/s41431-018-0121-7
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0121-7
This article is cited by
-
A genotype-first analysis in a cohort of Mullerian anomaly
Journal of Human Genetics (2022)
-
Biallelic ZNF407 mutations in a neurodevelopmental disorder with ID, short stature and variable microcephaly, hypotonia, ocular anomalies and facial dysmorphism
Journal of Human Genetics (2020)