Abstract
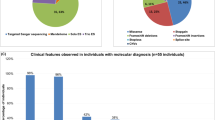
Hirschsprung disease (HSCR) is a complex birth defect characterized by the lack of ganglion cells along a variable length of the distal intestine. A large proportion of HSCR patients remain genetically unexplained. We applied whole-genome sequencing (WGS) on 9 trios where the probands are sporadically affected with the most severe form of the disorder and harbor no coding sequence variants affecting the function of known HSCR genes. We found de novo protein-altering variants in three intolerant to change genes—CCT2, VASH1, and CYP26A1—for which a plausible link with the enteric nervous system (ENS) exists. De novo single-nucleotide and indel variants were present in introns and non-coding neighboring regions of ENS-related genes, including NRG1 and ERBB4. Joint analysis with those inherited rare variants found under recessive and/or digenic models revealed both patient-unique and shared genetic features where rare variants were found to be enriched in the extracellular matrix–receptor (ECM–receptor) pathway (p = 3.4 × 10−11). Delineation of the genetic profile of each patient might help finding common grounds that could lead to the discovery of shared molecules that could be used as drug targets for the currently ongoing cell therapy effort which aims at providing an alternative to the surgical treatment.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Amiel J, Sproat-Emison E, Garcia-Barcelo M, et al. Hirschsprung disease, associated syndromes and genetics: a review. J Med Genet. 2008;45:1–14.
Emison ES, Garcia-Barcelo M, Grice EA, et al. Differential contributions of rare and common, coding and noncoding Ret mutations to multifactorial Hirschsprung disease liability. Am J Hum Genet. 2010;87:60–74.
Garcia-Barcelo MM, Sham MH, Lui VC, et al. Chinese patients with sporadic Hirschsprung’s disease are predominantly represented by a single RET haplotype. J Med Genet. 2003;40:e122.
Tang CS, Ngan ES, Tang WK et al. Mutations in the NRG1 gene are associated with Hirschsprung disease. Hum Genet. 2011;131:67–76.
Tang CS, Cheng G, So MT, et al. Genome-wide copy number analysis uncovers a new HSCR gene: NRG3. PLoS Genet. 2012;8:e1002687.
Jiang Q, Arnold S, Heanue T, et al. Functional loss of semaphorin 3C and/or semaphorin 3D and their epistatic interaction with ret are critical to hirschsprung disease liability. Am J Hum Genet. 2015;96:581–96.
Garcia-Barcelo MM, Tang CS, Ngan ES, et al. Genome-wide association study identifies NRG1 as a susceptibility locus for Hirschsprung’s disease. Proc Natl Acad Sci USA. 2009;106:2694–9.
Emison ES, McCallion AS, Kashuk CS, et al. A common sex-dependent mutation in a RET enhancer underlies Hirschsprung disease risk. Nature. 2005;434:857–63.
Fattahi F, Steinbeck JA, Kriks S, et al. Deriving human ENS lineages for cell therapy and drug discovery in Hirschsprung disease. Nature. 2016;531:105–9.
Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60.
McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
Li MX, Li J, Li MLJ et al. Robust and rapid algorithms facilitate large-scale whole genome sequencing downstream analysis in an integrative framework. Nucleic Acids Res. 2017; 45 e75.
Abyzov A, Urban AE, Snyder M, Gerstein M. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 2011;21:974–84.
Liang Y, Qiu K, Liao B, et al. Seeksv: an accurate tool for somatic structural variation and virus integration detection. Bioinformatics. 2017;33:184–91.
Rausch T, Zichner T, Schlattl A, Stutz AM, Benes V, Korbel JO. DELLY: structural variant discovery by integrated paired-end and split-read analysis. Bioinformatics. 2012;28:i333–i339.
Layer RM, Chiang C, Quinlan AR, Hall IM: LUMPY: a probabilistic framework for structural variant discovery. Genome Biol. 2014; 15 R84.
Quinlan AR. BEDTools: the Swiss-army tool for genome feature analysis. Curr Protoc Bioinformatics. 2014;47:11 12 11-34.
Yang H, Wang K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat Protoc. 2015;10:1556–66.
MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The Database of Genomic Variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992.
Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368.
Vlasblom J, Zuberi K, Rodriguez H, et al. Novel function discovery with GeneMANIA: a new integrated resource for gene function prediction in Escherichia coli. Bioinformatics. 2015;31:306–10.
Acuna-Hidalgo R, Veltman JA, Hoischen A. New insights into the generation and role of de novo mutations in health and disease. Genome Biol. 2016;17:241.
Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7.
Sato Y, Heuckeroth RO. Retinoic acid regulates murine enteric nervous system precursor proliferation, enhances neuronal precursor differentiation, and reduces neurite growth in vitro. Dev Biol. 2008;320:185–98.
Simkin JE, Zhang D, Rollo BN, Newgreen DF. Retinoic acid upregulates ret and induces chain migration and population expansion in vagal neural crest cells to colonise the embryonic gut. PLoS ONE. 2013;8:e64077.
Gui H, Schriemer D, Cheng WW, et al. Whole exome sequencing coupled with unbiased functional analysis reveals new Hirschsprung disease genes. Genome Biol. 2017;18:48.
Lai FP, Lau ST, Wong JK et al. Correction of Hirschsprung-associated Mutations in Human Induced Pluripotent Stem Cells, via CRISPR/Cas9, Restores Neural Crest Cell Function. Gastroenterology 2017;153:139–153.
Gershon MD, Ratcliffe EM. Developmental biology of the enteric nervous system: pathogenesis of Hirschsprung’s disease and other congenital dysmotilities. Semin Pediatr Surg. 2004;13:224–35.
Jamuar SS, Schmitz-Abe K, D’Gama AM, et al. Biallelic mutations in human DCC cause developmental split-brain syndrome. Nat Genet. 2017;49:606–12.
Marsh AP, Heron D, Edwards TJ, et al. Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nat Genet. 2017;49:511–4.
Sayed M, al-Alaiyan S. Agenesis of corpus callosum, hypertrophic pyloric stenosis and Hirschsprung disease: coincidence or common etiology? Neuropediatrics. 1996;27:204–6.
Pietri T, Eder O, Blanche M, Thiery JP, Dufour S. The human tissue plasminogen activator-Cre mouse: a new tool for targeting specifically neural crest cells and their derivatives in vivo. Dev Biol. 2003;259:176–87.
Smolen GA, Schott BJ, Stewart RA, et al. A Rap GTPase interactor, RADIL, mediates migration of neural crest precursors. Genes Dev. 2007;21:2131–6.
Sasselli V, Pachnis V, Burns AJ. The enteric nervous system. Dev Biol. 2012;366:64–73.
Soret R, Mennetrey M, Bergeron KF, et al. A collagen VI-dependent pathogenic mechanism for Hirschsprung’s disease. J Clin Invest. 2015;125:4483–96.
Zhang K, Wong P, Zhang L, et al. A Notch1-neuregulin1 autocrine signaling loop contributes to melanoma growth. Oncogene. 2012;31:4609–18.
Nagy N, Goldstein AM: Enteric nervous system development: a crest cell’s journey from neural tube to colon. Semin Cell Dev Biol. 2017;66:94–106.
Watanabe Y, Stanchina L, Lecerf L, et al. Differentiation of mouse enteric nervous system progenitor cells is controlled by endothelin 3 and requires regulation of Ednrb by SOX10 and ZEB2. Gastroenterology. 2017;152:1139–50. e1134
Brancati F, Dallapiccola B, Valente EM. Joubert Syndrome and related disorders. Orphanet J Rare Dis. 2010;5:20.
Waters AM, Beales PL. Ciliopathies: an expanding disease spectrum. Pediatr Nephrol. 2011;26:1039–56.
Wieczorek D, Krause M, Majewski F, et al. Unexpected high frequency of de novo unbalanced translocations in patients with Wolf-Hirschhorn syndrome (WHS). J Med Genet. 2000;37:798–804.
Garavelli L, Mainardi PC. Mowat-Wilson syndrome. Orphanet J Rare Dis. 2007;2:42.
Konopka G, Friedrich T, Davis-Turak J, et al. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–17.
Shi L. Dock protein family in brain development and neurological disease. Commun Integr Biol. 2013;6:e26839.
Samocha KE, Robinson EB, Sanders SJ, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46:944–50.
MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–76.
Acknowledgements
We are grateful to the numerous patients, their families and referring physicians that have participated in these studies in our laboratories, and the numerous members of our laboratories for their valuable contributions over many years.
Funding
This work was supported by: Theme-based Research scheme T12C-714/14-R to P.T., Human Medical Research Fund (HMRF) 02131866 and 01121516 to M.M.G.B and 0451966 to C.S.T. and Seed Fund for Basic Research of the University of Hong Kong 201606159005 to C.S.T.
Author information
Authors and Affiliations
Contributions
M.-M.G.-B., P.C.S., and P.K.T. conceived the experiments. C.S.T., X.Z. and M.-M.G.-B. wrote the article. P.C.S. and S.S.C. contributed to critical revision of the article. C.S.T. and X.Z. did the statistical analysis and data interpretation. W.Y.L. and J.S.H. assisted the analysis. M.T.S. did the genetic sample preparation and Sanger sequencing. E.S.W.N. provided the expression data. P.K.T., M.Y. and N.D.N. recruit and clinically characterized the patients.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The study was approved by the institutional review board of The University of Hong Kong together with the Hospital Authority (IRB: UW 06-349T/1374).
Informed consent
All participant information was anonymized and unique ID codes were presented in this manuscript. Publication of the de-identified results from all consenting participants was approved.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Tang, C.S., Zhuang, X., Lam, WY. et al. Uncovering the genetic lesions underlying the most severe form of Hirschsprung disease by whole-genome sequencing. Eur J Hum Genet 26, 818–826 (2018). https://doi.org/10.1038/s41431-018-0129-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0129-z
This article is cited by
-
Compound heterozygous mutations in a mouse model of Leber congenital amaurosis reveal the role of CCT2 in photoreceptor maintenance
Communications Biology (2024)
-
Genetics of Hirschsprung’s disease
Pediatric Surgery International (2023)
-
Stem cell-based therapy for hirschsprung disease, do we have the guts to treat?
Gene Therapy (2022)
-
“Too much guts and not enough brains”: (epi)genetic mechanisms and future therapies of Hirschsprung disease — a review
Clinical Epigenetics (2019)