Abstract
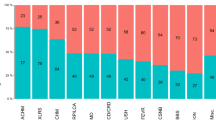
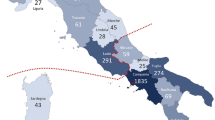
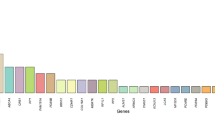
Inherited retinal diseases (IRDs) are heterogeneous phenotypes caused by variants in a large number of genes. Disease prevalence and the frequency of carriers in the general population have been estimated in only a few studies, but are largely unknown. To this end, we developed two parallel methods to calculate carrier frequency for mutations causing autosomal-recessive (AR) IRDs in the Israeli population. We created an SQL database containing information on 178 genes from gnomAD (including genotyping of 5706 Ashkenazi Jewish (AJ) individuals) and our cohort of >2000 families with IRDs. Carrier frequency for IRD variants and genes was calculated based on allele frequency values and the Hardy–Weinberg (HW) equation. We identified 399 IRD-causing variants in 111 genes in Israeli patients and AJ controls. For the AJ subpopulation, gnomAD and HW-based regression analysis showed high correlation, therefore allowing one to use HW-based data as a reliable estimate of carrier frequency. Overall, carrier frequency per subpopulation ranges from 1/2.2 to 1/9.6 individuals, with the highest value obtained for the Arab-Muslim subpopulation in Jerusalem reaching an extremely high carrier rate of 44.7%. Carrier frequency per gene ranges from 1/31 to 1/11994 individuals. We estimate the total carrier frequency for AR-IRD mutations in the Israeli population as over 30%, a relatively high carrier frequency with marked variability among subpopulations. Therefore, these data are highly important for more reliable genetic counseling and genetic screening. Our method can be adapted to study other populations, either based on allele frequency data or cohort of patients.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Chong JX, Ouwenga R, Anderson RL, Waggoner DJ, Ober C. A population-based study of autosomal-recessive disease-causing mutations in a founder population. Am J Hum Genet. 2012;91:608–20.
Gao Z, Waggoner D, Stephens M, Ober C, Przeworski M. An estimate of the average number of recessive lethal mutations carried by humans. Genetics. 2015;199:1243–54.
Macarthur DG, Balasubramanian S, Frankish A, et al. A systematic survey of loss-of-function variants in human protein-coding genes. 2012;335:823–8.
Xue Y, Chen Y, Ayub Q, et al. Deleterious- and disease-allele prevalence in healthy individuals: insights from current predictions, mutation databases, and population-scale resequencing. Am J Hum Genet. 2012;91:1022–32.
Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368:1795–809.
Sharon D, Banin E. Nonsyndromic retinitis pigmentosa is highly prevalent in the Jerusalem region with a high frequency of founder mutations. Mol Vis. 2015;21:783–92.
Nishiguchi KM, Rivolta C. Genes associated with retinitis pigmentosa and allied diseases are frequently mutated in the general population. PLoS ONE. 2012;7:5–7.
Rivolta C, Sharon D, DeAngelis MM, Dryja TP. Retinitis pigmentosa and allied diseases: numerous diseases, genes, and inheritance patterns. Hum Mol Genet. 2002;11:1219–27.
Lek M, Karczewski KJ, Minikel EV, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
Landrum MJ, Lee JM, Benson M. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44:D862–8.
Cremers FPM, Dunnen JT, den, Ajmal M, et al. Comprehensive registration of DNA sequence variants associated with inherited retinal diseases in Leiden Open Variation Databases. Hum Mutat. 2014;35:147–8.
Cornelis SS, Bax NM, Zernant J, et al. In silico functional meta-analysis of 5,962 ABCA4 variants in 3,928 retinal dystrophy cases. Hum Mutat. 2017;38:400–8.
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–63.
Stenson PD, Mort M, Ball EV, Shaw K, Phillips AD, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9.
Sleat DE, Gedvilaite E, Zhang Y, Lobel P, Xing J. Analysis of large-scale whole exome sequencing data to determine the prevalence of genetically-distinct forms of neuronal ceroid lipofuscinosis. Gene. 2016;593:284–91.
Auslender N, Sharon D, Abbasi AH, Garzozi HJ, Banin E, Ben-Yosef T. A common founder mutation of CERKL underlies autosomal recessive retinal degeneration with early macular involvement among Yemenite Jews. Invest Ophthalmol Vis Sci. 2007;48:5431–8.
Zelinger L, Greenberg A, Kohl S, Banin E, Sharon D. An ancient autosomal haplotype bearing a rare achromatopsia-causing founder mutation is shared among Arab Muslims and Oriental Jews. Hum Genet. 2010;128:261–7.
Khateb S, Hanany M, Khalailah A, et al. Identification of genomic deletions causing inherited retinal degenerations by coverage analysis of whole exome sequencing data. J Med Genet. 2016;53:600–7.
Van Cauwenbergh C, Van Schil K, Cannoodt R, et al. arrEYE: a customized platform for high-resolution copy number analysis of coding and noncoding regions of known and candidate retinal dystrophy genes and retinal noncoding RNAs. Genet Med. 2017;19:457–66.
Hardy GH. Mendelian proportions in a mixed population. Science. 1908;28:49–50.
Weinberg W. Über den Nachweis der Vererbung beim Menschen. Jahresh Des Ver für Vaterl Nat Württemberg. 1908;64:368–82.
Guymer RH, Heon E, Lotery AJ, et al. Variation of codons 1961 and 2177 of the Stargardt disease gene is not associated with age-related macular degeneration. Arch Ophthalmol. 2001;119:745–51.
Schulz HL, Grassmann F, Kellner U, et al. Mutation spectrum of the ABCA4 gene in 335 Stargardt disease patients from a multicenter german cohort—impact of selected deep intronic variants and common SNPs. Invest Ophthalmol Vis Sci. 2017;58:394–403.
Bax NM, Sangermano R, Roosing S, et al. Heterozygous deep-intronic variants and deletions in ABCA4 in persons with retinal dystrophies and one exonic ABCA4 variant. Hum Mutat. 2015;36:43–7.
Braun TA, Mullins RF, Wagner AH, et al. Non-exomic and synonymous variants in ABCA4 are an important cause of Stargardt disease. Hum Mol Genet. 2013;22:5136–45.
Bauwens M, De Zaeytijd J, Weisschuh N, et al. An augmented ABCA4 screen targeting noncoding regions reveals a deep intronic founder variant in Belgian Stargardt Patients. Hum Mutat. 2015;36:39–42.
Nishiguchi KM, Sandberg MA, Kooijman AC, et al. Defects in RGS9 or its anchor protein R9AP in patients with slow photoreceptor deactivation. Nature. 2004;427:75–8.
Acknowledgements
This study was funded by the Israeli Ministry of Health (grant numbers 3-10999 to D.S. and T.B.Y. and 3-12583 to H.N., E.P., L.G., E.B., T.B.Y., and D.S.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hanany, M., Allon, G., Kimchi, A. et al. Carrier frequency analysis of mutations causing autosomal-recessive-inherited retinal diseases in the Israeli population. Eur J Hum Genet 26, 1159–1166 (2018). https://doi.org/10.1038/s41431-018-0152-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-018-0152-0