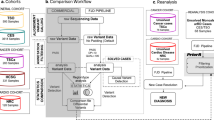
Abstract
Clinical exome sequencing (CES) is increasingly being utilized; however, a large proportion of patients remain undiagnosed, creating a need for a systematic approach to increase the diagnostic yield. We have reanalyzed CES data for a clinically heterogeneous cohort of 102 probands with likely Mendelian conditions, including 74 negative cases and 28 cases with candidate variants, but reanalysis requested by clinicians. Reanalysis was performed by an interdisciplinary team using a validated custom-built pipeline, “Variant Explorer Pipeline” (VExP). This reanalysis approach and results were compared with existing literature. Reanalysis of candidate variants from CES in 28 cases revealed 1 interpretation that needed to be reclassified. A confirmed or potential genetic diagnosis was identified in 24 of 75 CES-negative/reclassified cases (32.0%), including variants in known disease-causing genes (n = 6) or candidate genes (n = 18). This yield was higher compared with similar studies demonstrating the utility of this approach. In summary, reanalysis of negative CES in a research setting enhances diagnostic yield by about a third. This study suggests the need for comprehensive, continued reanalysis of exome data when molecular diagnosis is elusive.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Stark Z, Schofield D, Alam K, Wilson W, Mupfeki N, Macciocca I, et al. Prospective comparison of the cost-effectiveness of clinical whole-exome sequencing with that of usual care overwhelmingly supports early use and reimbursement. Genet Med. 2017;19:867–74.
Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–6.
Xue Y, Ankala A, Wilcox WR, Hegde MR. Solving the molecular diagnostic testing conundrum for Mendelian disorders in the era of next-generation sequencing: single-gene, gene panel, or exome/genome sequencing. Genet Med. 2015;17:444–51.
Bamshad MJ, Ng SB, Bigham AW, Tabor HK, Emond MJ, Nickerson DA, et al. Exome sequencing as a tool for Mendelian disease gene discovery. Nat Rev Genet. 2011;12:745–55.
Wenger AM, Guturu H, Bernstein JA, Bejerano G. Systematic reanalysis of clinical exome data yields additional diagnoses: implications for providers. Genet Med. 2017;19:209–14.
Pena LD, Jiang Y-H, Schoch K, Spillmann RC, Walley N, Stong N, et al. Looking beyond the exome: a phenotype-first approach to molecular diagnostic resolution in rare and undiagnosed diseases. Genet Med. 2018;20:464–9.
Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, et al. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–7.
Farwell KD, Shahmirzadi L, El-Khechen D, Powis Z, Chao EC, Davis BT, et al. Enhanced utility of family-centered diagnostic exome sequencing with inheritance model–based analysis: results from 500 unselected families with undiagnosed genetic conditions. Genet Med. 2015;17:578–86.
Bone WP, Washington NL, Buske OJ, Adams DR, Davis J, Draper D, et al. Computational evaluation of exome sequence data using human and model organism phenotypes improves diagnostic efficiency. Genet Med. 2016;18:608–17.
Eldomery MK, Coban-Akdemir Z, Harel T, Rosenfeld JA, Gambin T, Stray-Pedersen A, et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Med. 2017;9:26.
Shashi V, Schoch K, Spillmann R, Cope H, Tan QK-G, Walley N, et al. A comprehensive iterative approach is highly effective in diagnosing individuals who are exome negative. Genet Med. 2019;21:161–72.
Hagman KDF, Shinde DN, Mroske C, Smith E, Radtke K, Shahmirzadi L, et al. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet Med. 2017;19:224–35.
Schmitz-Abe K, Ciesielski SJ, Schmidt PJ, Campagna DR, Rahimov F, Schilke BA, et al. Congenital sideroblastic anemia due to mutations in the mitochondrial HSP70 homologue HSPA9. Blood. 2015;126:2734–8.
Jamuar SS, Schmitz-Abe K, D’Gama AM, Drottar M, Chan W-M, Peeva M, et al. Biallelic mutations in human DCC cause developmental split-brain syndrome. Nat Genet. 2017;49:606–12.
Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, Brudno M, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat. 2015;36:915–21.
Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–24.
Küry S, van Woerden GM, Besnard T, Onori MP, Latypova X, Towne MC, et al. De novo mutations in protein kinase genes CAMK2A and CAMK2B cause intellectual disability. Am J Hum Genet. 2017;101:768–88.
Hamdan FF, Myers CT, Cossette P, Lemay P, Spiegelman D, Laporte AD, et al. High rate of recurrent de novo mutations in developmental and epileptic encephalopathies. Am J Hum Genet. 2017;101:664–85.
Walters GB, Gustafsson O, Sveinbjornsson G, Eiriksdottir VK, Agustsdottir AB, Jonsdottir GA, et al. MAP1B mutations cause intellectual disability and extensive white matter deficit. Nat Commun. 2018;9:3456.
Nambot S, Thevenon J, Kuentz P, Duffourd Y, Tisserant E, Bruel A-L, et al. Clinical whole-exome sequencing for the diagnosis of rare disorders with congenital anomalies and/or intellectual disability: substantial interest of prospective annual reanalysis. Genet Med. 2018;20:645–54.
Wright CF, McRae JF, Clayton S, Gallone G, Aitken S, FitzGerald TW, et al. Making new genetic diagnoses with old data: iterative reanalysis and reporting from genome-wide data in 1,133 families with developmental disorders. Genet Med. 2018;20:1216–23.
Al‐Nabhani M, Al‐Rashdi S, Al‐Murshedi F, Al‐Kindi A, Al‐Thihli K, Al‐Saegh A, et al. Re‐analysis of exome sequencing data of intellectual disability samples: yields and benefits. Clin Genet. 2018;94:495–501.
Shamseldin HE, Maddirevula S, Faqeih E, Ibrahim N, Hashem M, Shaheen R, et al. Increasing the sensitivity of clinical exome sequencing through improved filtration strategy. Genet Med. 2017;19:593–8.
Mersch J, Brown N, Pirzadeh-Miller S, Mundt E, Cox HC, Brown K, et al. Prevalence of variant reclassification following hereditary cancer genetic testing. JAMA. 2018;320:1266–74.
Bergant G, Maver A, Lovrecic L, Čuturilo G, Hodzic A, Peterlin B. Comprehensive use of extended exome analysis improves diagnostic yield in rare disease: a retrospective survey in 1,059 cases. Genet Med. 2018;20:303–12.
Deltas C. Digenic inheritance and genetic modifiers. Clin Genet. 2018;93:429–38.
Li H. Toward better understanding of artifacts in variant calling from high-coverage samples. Bioinformatics. 2014;30:2843–51.
Shirley MD, Tang H, Gallione CJ, Baugher JD, Frelin LP, Cohen B, et al. Sturge–Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368:1971–9.
Tankard RM, Delatycki MB, Lockhart PJ, Bahlo M. Detecting known repeat expansions with standard protocol next generation sequencing, towards developing a single screening test for neurological repeat expansion disorders. Am J Hum Genet. 2018;103:858–73.
Wenstrom KD. Fragile X and other trinucleotide repeat diseases. Obstet Gynecol Clin North Am. 2002;29:367–88.
Gajecka M. Unrevealed mosaicism in the next-generation sequencing era. Mol Genet Genomics. 2016;291:513–30.
Lionel AC, Costain G, Monfared N, Walker S, Reuter MS, Hosseini SM, et al. Improved diagnostic yield compared with targeted gene sequencing panels suggests a role for whole-genome sequencing as a first-tier genetic test. Genet Med. 2018;20:435–43.
Byron SA, Van Keuren-Jensen KR, Engelthaler DM, Carpten JD, Craig DW. Translating RNA sequencing into clinical diagnostics: opportunities and challenges. Nat Rev Genet. 2016;17:257–71.
Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, et al. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43:838–46.
Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903.
Acknowledgements
We thank all the referring physicians and caregivers, and especially the patients and families for their participation in this research. We also thank Heather Paterson, Tina Truong, Grace VanNoy, and Clair McHugh for assistance with patient enrollment and sample and data collection. The Manton Center Gene Discovery Core is supported by a generous gift from The Manton Foundation. This work was also supported by grants NIAMS R01AR068429, NICHD/NHGRI U19HD077671, NICHD R01HD075802, and NICHD K12HD052896, and by the resources of the IDDRC Molecular Genetics Core funded by U54HD090255, from the US National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Schmitz-Abe, K., Li, Q., Rosen, S.M. et al. Unique bioinformatic approach and comprehensive reanalysis improve diagnostic yield of clinical exomes. Eur J Hum Genet 27, 1398–1405 (2019). https://doi.org/10.1038/s41431-019-0401-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0401-x
This article is cited by
-
Implementation of multi-omics in diagnosis of pediatric rare diseases
Pediatric Research (2025)
-
Novel MAP1B loss-of-function variant associated with periventricular nodular heterotopia 9 and literature review on genotype-phenotype associations of MAP1B
Neurological Sciences (2025)
-
Systematic reanalysis of genomic data by diagnostic laboratories: a scoping review of ethical, economic, legal and (psycho)social implications
European Journal of Human Genetics (2024)
-
Variants in mitochondrial disease genes are common causes of inherited peripheral neuropathies
Journal of Neurology (2024)
-
Combining a prioritization strategy and functional studies nominates 5’UTR variants underlying inherited retinal disease
Genome Medicine (2024)