Abstract
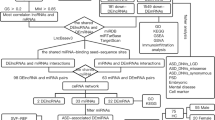
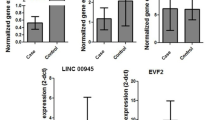
We identified a 14q21.2 microdeletion in a 16-year-old boy with autism spectrum disorder (ASD), IQ in the lower part of normal range but high-functioning memory skills. The deletion affects a gene desert, and the non-deleted gene closest to the microdeletion boundaries is LRFN5, which encodes a protein involved in synaptic plasticity and implicated in neuro-psychiatric disorders. LRFN5 expression was significantly decreased in the proband’s skin fibroblasts. The deleted region includes the pseudogene chr14.232.a, which is transcribed into a long non-coding RNA (lncLRFN5-10), whose levels were also significantly reduced in the proband’s fibroblasts compared to controls. Transfection of the patient’s fibroblasts with a plasmid expressing chr14.232.a significantly increased LRFN5 expression, while siRNA targeting chr14.232.a-derived lncLRFN5-10 reduced LRFN5 levels. In summary, we report on an individual with ASD carrying a microdeletion encompassing the pseudogene chr14.232.a encoding for lncLRFN5-10, which was found to affect the expression levels of the nearby, non-deleted LRFN5. This case illustrates the potential role of long non-coding RNAs in regulating expression of neighbouring genes with a functional role in ASD pathogenesis.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Christensen DL, Baio J, Van Naarden Braun K, Bilder D, Charles J, Constantino JN, et al. Prevalence and characteristics of autism spectrum disorder among children aged 8 years—autism and developmental disabilities monitoring network, 11 sites, United States, 2012. MMWR Surveill Summ. 2016;65:1–23.
Tammimies K, Marshall CR, Walker S, Kaur G, Thiruvahindrapuram B, Lionel AC, et al. Molecular diagnostic yield of chromosomal microarray analysis and whole-exome sequencing in children with autism spectrum disorder. JAMA. 2015;314:895–903.
Pereanu W, Larsen EC, Das I, Estevez MA, Sarkar AA, Spring-Pearson S, et al. AutDB: a platform to decode the genetic architecture of autism. Nucleic Acids Res. 2018;46:D1049–D1054.
Short PJ, McRae JF, Gallone G, Sifrim A, Won H, Geschwind DH, et al. De novo mutations in regulatory elements in neurodevelopmental disorders. Nature. 2018;555:611–6.
Choi Y, Nam J, Whitcomb DJ, Song YS, Kim D, Jeon S, et al. SALM5 trans-synaptically interacts with LAR-RPTPs in a splicing-dependent manner to regulate synapse development. Sci Rep. 2016;6:26676.
Goto-Ito S, Yamagata A, Sato Y, Uemura T, Shiroshima T, Maeda A, et al. Structural basis of trans-synaptic interactions between PTPdelta and SALMs for inducing synapse formation. Nat Commun. 2018;9:269.
Spielmann M, Lupianez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19:453–67.
Volders PJ, Verheggen K, Menschaert G, Vandepoele K, Martens L, Vandesompele J, et al. An update on LNCipedia: a database for annotated human lncRNA sequences. Nucleic Acids Res. 2015;43:4363–4.
Lin Z, Liu J, Ding H, Xu F, Liu H. Structural basis of SALM5-induced PTPdelta dimerization for synaptic differentiation. Nat Commun. 2018;9:268.
Xu B, Woodroffe A, Rodriguez-Murillo L, Roos JL, van Rensburg EJ, Abecasis GR, et al. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc Natl Acad Sci USA. 2009;106:16746–51.
Mikhail FM, Lose EJ, Robin NH, Descartes MD, Rutledge KD, Rutledge SL, et al. Clinically relevant single gene or intragenic deletions encompassing critical neurodevelopmental genes in patients with developmental delay, mental retardation, and/or autism spectrum disorders. Am J Med Genet Part A. 2011;155:2386–96.
Otter M, Schrander-Stumpel CT, Curfs LM. Triple X syndrome: a review of the literature. Eur J Hum Genet. 2010;18:265–71.
de Bruijn DRH, van Dijk AHA, Pfundt R, Hoischen A, Merkx GFM, Gradek GA, et al. Severe progressive autism associated with two de novo hanges: a 2.6-Mb 2q31.1 deletion and a balanced t(14;21)(q21.1; p11.2) translocation with long-range epigenetic silencing of LRFN5 expression. Mol Syndromol. 2010;1:46–57.
Farwell Hagman KD, Shinde DN, Mroske C, Smith E, Radtke K, Shahmirzadi L, et al. Candidate-gene criteria for clinical reporting: diagnostic exome sequencing identifies altered candidate genes among 8% of patients with undiagnosed diseases. Genet Med. 2017;19:224–35.
Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, et al. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs). Mol Autism. 2013;4:36.
Turner TN, Yi Q, Krumm N, Huddleston J, Hoekzema K, FS HA, et al. Denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2017;45:D804–D811.
Knauss JL, Sun T. Regulatory mechanisms of long noncoding RNAs in vertebrate central nervous system development and function. Neuroscience. 2013;235:200–14.
Barry G. Integrating the roles of long and small non-coding RNA in brain function and disease. Mol Psychiatry. 2014;19:410–6.
Parikshak NN, Swarup V, Belgard TG, Irimia M, Ramaswami G, Gandal MJ, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540:423–7.
Briggs JA, Wolvetang EJ, Mattick JS, Rinn JL, Barry G. Mechanisms of long non-coding RNAs in mammalian nervous system development, plasticity, disease, and evolution. Neuron. 2015;88:861–77.
Wahlestedt C. Targeting long non-coding RNA to therapeutically upregulate gene expression. Nat Rev Drug Discov. 2013;12:433–46.
Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–83.
Poot M. A candidate gene association study further corroborates involvement of contactin genes in autism. Mol Syndromol. 2014;5:229–35.
Pizzo L, Jensen M, Polyak A, Rosenfeld JA, Mannik K, Krishnan A, et al. Rare variants in the genetic background modulate cognitive and developmental phenotypes in individuals carrying disease-associated variants. Genet Med. 2019;21:816–25.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Cappuccio, G., Attanasio, S., Alagia, M. et al. Microdeletion of pseudogene chr14.232.a affects LRFN5 expression in cells of a patient with autism spectrum disorder. Eur J Hum Genet 27, 1475–1480 (2019). https://doi.org/10.1038/s41431-019-0430-5
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0430-5
This article is cited by
-
Screening and function analysis of key genes and signaling pathways in Alzheimer’s disease via next generation sequencing data analysis and bioinformatics approaches
Genome Instability & Disease (2025)
-
Two polygenic mouse models of major depressive disorders identify TMEM161B as a potential biomarker of disease in humans
Neuropsychopharmacology (2024)
-
Dissecting diagnostic heterogeneity in depression by integrating neuroimaging and genetics
Neuropsychopharmacology (2021)