Abstract
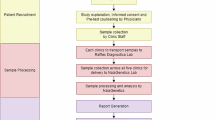
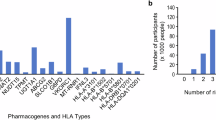
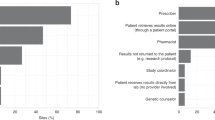
Despite the nationwide availability of pharmacogenomic (PGx) guidelines in electronic medication surveillance systems in The Netherlands, PGx guided prescribing is still uncommon in primary care. We set out to investigate the adoption of pharmacist initiated PGx testing in primary care. Community pharmacists were offered a free PGx test covering 40 variants in 8 genes to test patients receiving an incident prescription (IRx) of a selection of 10 drugs. Results of the PGx test along with predicted phenotypes and a therapeutic recommendation based on the Dutch Pharmacogenetics Working Group (DPWG) guidelines were transferred to the pharmacist and physician. Adoption was defined as the percentage of eligible patients that received genotyping. From November 2014–July 2016, 200 patients were included with an adoption of 18.0%. Of the included patients 57.5% received an IRx for atorvastatin, 14.5% started with simvastatin and 28.0% received an IRx for amitriptyline, (es)citalopram, nortriptyline, or venlafaxine. 90% of the patients carried at least one actionable PGx test result in the selected PGx-panel. In 31.0% of the incident prescriptions a combination between a drug with a known gene-drug interaction and an actionable genotype was present and a therapeutic recommendation was provided. The provided recommendations were accepted by the clinicians in 88.7% of the patients. Pharmacist initiated implementation of PGx in primary care is feasible, and the frequency of actionable gene-drug interactions for the selected drugs is high.
Similar content being viewed by others
Log in or create a free account to read this content
Gain free access to this article, as well as selected content from this journal and more on nature.com
or
References
Swen JJ, Huizinga TW, Gelderblom H, de Vries EG, Assendelft WJ, Kirchheiner J, et al. Translating pharmacogenomics: challenges on the road to the clinic. PLoS Med. 2007;4:e209.
Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther. 2011;89:464–7.
Swen JJ, Wilting I, de Goede AL, Grandia L, Mulder H, Touw DJ, et al. Pharmacogenetics: from bench to byte. Clin Pharmacol Ther. 2008;83:781–7.
Swen JJ, Nijenhuis M, de Boer A, Grandia L, Maitland-van der Zee AH, Mulder H, et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther. 2011;89:662–73.
Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. Am J Med Genet Part C Semin Med Genet. 2014;166C:45–55.
van Driest SL, Shi Y, Bowton EA, Schildcrout JS, Peterson JF, Pulley J, et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther. 2014;95:423–31.
O’Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clin Pharmacol Ther. 2012;92:446–9.
O’Donnell PH, Danahey K, Jacobs M, Wadhwa NR, Yuen S, Bush A, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care—initial results of the University of Chicago “1200 Patients Project”. Am J Med Genet Part C Semin Med Genet. 2014;166C:68–75.
Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc. 2014;89:25–33.
Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14:723–6.
Dunnenberger HM, Crews KR, Hoffman JM, Caudle KE, Broeckel U, Howard SC, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharm Toxicol. 2015;55:89–106.
Bank PC, Swen JJ, Guchelaar HJ. A nationwide survey of pharmacists’ perception of pharmacogenetics in the context of a clinical decision support system containing pharmacogenetics dosing recommendations. Pharmacogenomics. 2017;18:215–25.
Kantor ED, Rehm CD, Haas JS, Chan AT, Giovannucci EL. Trends in Prescription Drug Use Among Adults in the United States From 1999-2012. J Am Med Assoc. 2015;314(17):1818–31.
Brixner D, Biltaji E, Bress A, Unni S, Ye X, Mamiya T, et al. The effect of pharmacogenetic profiling with a clinical decision support tool on healthcare resource utilization and estimated costs in the elderly exposed to polypharmacy. J Med Econ. 2016;19:213–28.
Elliott LS, Henderson JC, Neradilek MB, Moyer NA, Ashcraft KC, Thirumaran RK. Clinical impact of pharmacogenetic profiling with a clinical decision support tool in polypharmacy home health patients: A prospective pilot randomized controlled trial. PLoS One. 2017;12:e0170905.
Finkelstein J, Friedman C, Hripcsak G, Cabrera M. Pharmacogenetic polymorphism as an independent risk factor for frequent hospitalizations in older adults with polypharmacy: a pilot study. Pharm Pers Med. 2016;9:107–16.
Perez V, Salavert A, Espadaler J, Tuson M, Saiz-Ruiz J, Saez-Navarro C, et al. Efficacy of prospective pharmacogenetic testing in the treatment of major depressive disorder: results of a randomized, double-blind clinical trial. BMC Psychiatry. 2017;17:250.
Swen JJ, van der Straaten T, Wessels JA, Bouvy ML, Vlassak EE, Assendelft WJ, et al. Feasibility of pharmacy-initiated pharmacogenetic screening for CYP2D6 and CYP2C19. Eur J Clin Pharmacol. 2012;68:363–70.
Dezentje VO, Opdam FL, Gelderblom H, Hartigh den J, Van der Straaten T, Vree R, et al. CYP2D6 genotype- and endoxifen-guided tamoxifen dose escalation increases endoxifen serum concentrations without increasing side effects. Breast Cancer Res Treat. 2015;153:583–90.
Lunenburg CA, van Staveren MC, Gelderblom H, Guchelaar HJ, Swen JJ. Evaluation of clinical implementation of prospective DPYD genotyping in 5-fluorouracil- or capecitabine-treated patients. Pharmacogenomics. 2016;17:721–9.
Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: the next generation in gene variant databases. Hum Mutat. 2011;32:557–63.
Boomsma DI, Wijmenga C, Slagboom EP, Swertz MA, Karssen LC, Abdellaoui A, et al. The Genome of the Netherlands: design, and project goals. Eur J Hum Genet: Ejhg. 2014;22:221–7.
Genome of the Netherlands Consortium. Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat Genet. 2014;46:818–25.
Francioli LC, Polak PP, Koren A, Menelaou A, Chun S, Renkens I, et al. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47:822–6.
Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91.
van der Wouden CH, Cambon-Thomsen A, Cecchin E, Cheung KC, Lucia Davila-Fajardo C, Deneer VH, et al. Implementing pharmacogenomics in Europe: design and implementation strategy of the ubiquitous pharmacogenomics consortium. Clin Pharmacol Ther. 2017;101:341–58.
Haga SB, O’Daniel JM, Tindall GM, Lipkus IR, Agans R. Survey of US public attitudes toward pharmacogenetic testing. Pharm J. 2012;12:197–204.
O’Daniel J, Lucas J, Deverka P, Ermentrout D, Silvey G, Lobach DF, et al. Factors influencing uptake of pharmacogenetic testing in a diverse patient population. Public Health Genom. 2010;13:48–54.
Acknowledgements
We would like to thank V. Rabiea for her assistance in the collection of the follow-up data. This study makes use of data generated by the Genome of the Netherlands (GoNl) Project and the Genome Aggregation Database (gnomAD). A full list of the investigators of the Genome of the Netherlands Project is available from http://www.nlgenome.nl. Funding for the GoNl project was provided by the Netherlands Organization for Scientific Research under award number 184021007, dated 9 July 2009 and made available as a Rainbow Project of the Biobanking and Biomolecular Research Infrastructure Netherlands (BBMRI-NL). The sequencing of samples included in the GoNl project was carried out in collaboration with the Beijing Institute for Genomics (BGI). Additionally, the authors would like to thank the Genome Aggregation Database (gnomAD) and the groups that provided exome and genome variant data to this resource. A full list of contributing groups can be found at https://gnomad.broadinstitute.org/about. This work was supported by unrestricted grants from The Royal Dutch Pharmacist’s Association, The Hague (KNMP), The Netherlands and Zorg & Zekerheid, Leiden, The Netherlands.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Bank, P.C.D., Swen, J.J., Schaap, R.D. et al. A pilot study of the implementation of pharmacogenomic pharmacist initiated pre-emptive testing in primary care. Eur J Hum Genet 27, 1532–1541 (2019). https://doi.org/10.1038/s41431-019-0454-x
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-019-0454-x
This article is cited by
-
Pharmacogenetic interventions to improve outcomes in patients with multimorbidity or prescribed polypharmacy: a systematic review
The Pharmacogenomics Journal (2022)
-
Clinical implementation of drug metabolizing gene-based therapeutic interventions worldwide
Human Genetics (2022)
-
Appraisal and development of evidence-based clinical decision support to enable perioperative pharmacogenomic application
The Pharmacogenomics Journal (2021)
-
Estimated nationwide impact of implementing a preemptive pharmacogenetic panel approach to guide drug prescribing in primary care in The Netherlands
BMC Medicine (2019)