Abstract
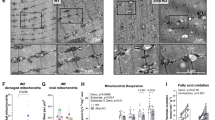
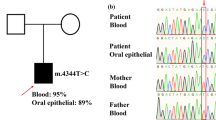
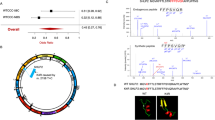
In a Dutch non-consanguineous patient having mitochondrial encephalomyopathy with complex I and complex IV deficiency, whole exome sequencing revealed two compound heterozygous variants in SLIRP. SLIRP gene encodes a stem-loop RNA-binding protein that regulates mitochondrial RNA expression and oxidative phosphorylation (OXPHOS). A frameshift and a deep-intronic splicing variant reduced the amount of functional wild-type SLIRP RNA to 5%. Consequently, in patient fibroblasts, MT-ND1, MT-ND6, and MT-CO1 expression was reduced. Lentiviral transduction of wild-type SLIRP cDNA in patient fibroblasts increased MT-ND1, MT-ND6, and MT-CO1 expression (2.5–7.2-fold), whereas mutant cDNAs did not. A fourfold decrease of citrate synthase versus total protein ratio in patient fibroblasts indicated that the resulting reduced mitochondrial mass caused the OXPHOS deficiency. Transduction with wild-type SLIRP cDNA led to a 2.4-fold increase of this ratio and partly restored OXPHOS activity. This confirmed causality of the SLIRP variants. In conclusion, we report SLIRP variants as a novel cause of mitochondrial encephalomyopathy with OXPHOS deficiency.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Russell OM, Gorman GS, Lightowlers RN, Turnbull DM. Mitochondrial diseases: hope for the future. Cell. 2020;181:168–88.
Rahman S. Mitochondrial disease in children. J Intern Med. 2020;287:609–33.
Thompson K, Collier JJ, Glasgow RIC, Robertson FM, Pyle A, Blakely EL, et al. Recent advances in understanding the molecular genetic basis of mitochondrial disease. J Inherit Metab Dis. 2020;43:36–50.
Almannai M, Alasmari A, Alqasmi A, Faqeih E, Al Mutairi F, Alotaibi M, et al. Expanding the phenotype of SLC25A42-associated mitochondrial encephalomyopathy. Clin Genet. 2018;93:1097–102.
Bruni F, Di Meo I, Bellacchio E, Webb BD, McFarland R, Chrzanowska-Lightowlers ZMA, et al. Clinical, biochemical, and genetic features associated with VARS2-related mitochondrial disease. Hum Mutat. 2018;39:563–78.
Gai X, Ghezzi D, Johnson MA, Biagosch CA, Shamseldin HE, Haack TB, et al. Mutations in FBXL4, encoding a mitochondrial protein, cause early-onset mitochondrial encephalomyopathy. Am J Hum Genet. 2013;93:482–95.
Baughman JM, Nilsson R, Gohil VM, Arlow DH, Gauhar Z, Mootha VK. A computational screen for regulators of oxidative phosphorylation implicates SLIRP in mitochondrial RNA homeostasis. PLoS Genet. 2009;5:e1000590.
Hatchell EC, Colley SM, Beveridge DJ, Epis MR, Stuart LM, Giles KM, et al. SLIRP, a small SRA binding protein, is a nuclear receptor corepressor. Mol Cell. 2006;22:657–68.
Lagouge M, Mourier A, Lee HJ, Spahr H, Wai T, Kukat C, et al. SLIRP Regulates the Rate of Mitochondrial Protein Synthesis and Protects LRPPRC from Degradation. PLoS Genet. 2015;11:e1005423.
Ruzzenente B, Metodiev MD, Wredenberg A, Bratic A, Park CB, Cámara Y, et al. LRPPRC is necessary for polyadenylation and coordination of translation of mitochondrial mRNAs. EMBO J. 2012;31:443–56.
Sasarman F, Brunel-Guitton C, Antonicka H, Wai T, Shoubridge EA, Consortium L. LRPPRC and SLIRP interact in a ribonucleoprotein complex that regulates posttranscriptional gene expression in mitochondria. Mol Biol Cell. 2010;21:1315–23.
Mootha VK, Lepage P, Miller K, Bunkenborg J, Reich M, Hjerrild M, et al. Identification of a gene causing human cytochrome c oxidase deficiency by integrative genomics. Proc Natl Acad Sci. 2003;100:605–10.
Spruijt L, Smeets HJ, Hendrickx A, Bettink-Remeijer MW, Maat-Kievit A, Schoonderwoerd KC, et al. A MELAS-Associated ND1 Mutation Causing Leber Hereditary Optic Neuropathy and Spastic Dystonia. Arch Neurol. 2007;64:890–3.
Theunissen TEJ, Nguyen M, Kamps R, Hendrickx AT, Sallevelt S, Gottschalk RWH, et al. Whole Exome Sequencing Is the Preferred Strategy to Identify the Genetic Defect in Patients With a Probable or Possible Mitochondrial Cause. Front Genet. 2018;9:400.
Nguyen M, Boesten I. Hellebrekers DMEI, Vanoevelen J, Kamps R, de Koning B, et al. Pathogenic CWF19L1 variants as a novel cause of autosomal recessive cerebellar ataxia and atrophy. Eur J Hum Genet. 2016;24:619–22.
Durand C, Roeth R, Dweep H, Vlatkovic I, Decker E, Schneider KU, et al. Alternative splicing and nonsense-mediated RNA decay contribute to the regulation of SHOX expression. PLoS ONE. 2011;6:e18115.
van den Bosch BJ, Gerards M, Sluiter W, Stegmann AP, Jongen EL, Hellebrekers DM, et al. Defective NDUFA9 as a novel cause of neonatally fatal complex I disease. J Med Genet. 2012;49:10–5.
Lehtonen JM, Auranen M, Darin N, Sofou K, Bindoff L, Hikmat O, et al. Diagnostic value of serum biomarkers FGF21 and GDF15 compared to muscle sample in mitochondrial disease. J Inherit Metab Dis. 2021;44:469–80.
Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics. 2011;10:M111.013284.
Beltrao P, Albanèse V, Kenner LR, Swaney DL, Burlingame A, Villén J, et al. Systematic functional prioritization of protein posttranslational modifications. Cell. 2012;150:413–25.
Stes E, Laga M, Walton A, Samyn N, Timmerman E, De Smet I, et al. A COFRADIC protocol to study protein ubiquitination. J Proteome Res. 2014;13:3107–13.
Chujo T, Ohira T, Sakaguchi Y, Goshima N, Nomura N, Nagao A, et al. LRPPRC/SLIRP suppresses PNPase-mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Res. 2012;40:8033–47.
Bruni F, Proctor-Kent Y, Lightowlers RN, Chrzanowska-Lightowlers ZM. Messenger RNA delivery to mitoribosomes—hints from a bacterial toxin. FEBS J. 2021;288:437–51.
Siira SJ, Spahr H, Shearwood AJ, Ruzzenente B, Larsson NG, Rackham O, et al. LRPPRC-mediated folding of the mitochondrial transcriptome. Nat Commun. 2017;8:1532.
Spahr H, Rozanska A, Li X, Atanassov I, Lightowlers RN, Chrzanowska-Lightowlers ZM, et al. SLIRP stabilizes LRPPRC via an RRM-PPR protein interface. Nucleic Acids Res. 2016;44:6868–82.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med: Off J Am Coll Med Genet. 2015;17:405–24.
Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, et al. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun. 2017;8:15824.
Acknowledgements
The authors are grateful to all participants for their involvement.
Funding
This study was supported by funding from China Scholarship Council (Grant Number: 201606310036) and Neuromusculair en Mitochondrieel Onderzoek (NeMO) foundation (Grant/Award Number: 19_P02). Part of this work has been supported by the Dutch Province of Limburg.
Author information
Authors and Affiliations
Contributions
LG, BPHE, MG contributed to the design of the research. LG wrote the manuscript. LG, BPHE, MG analysed the WES data. LG, BPHE, IMGMH performed the cellular experiments. MV, SCEHS, DMEIH provided the clinical data. EHJ, FSN performed the OXPHOS measurements. FHJT, HJMS, MG were involved in planning and supervised the work. All authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Follow-up investigations to confirm the genetic diagnosis were performed in accordance with the regulations of the local ethical committee of Maastricht University Medical Centre.
Informed consent
Subjects gave written informed consent for WES analysis in accordance with the Declaration of Helsinki as part of their clinical/molecular genetics diagnosis at Maastricht University Medical Centre.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Guo, L., Engelen, B.P.H., Hemel, I.M.G.M. et al. Pathogenic SLIRP variants as a novel cause of autosomal recessive mitochondrial encephalomyopathy with complex I and IV deficiency. Eur J Hum Genet 29, 1789–1795 (2021). https://doi.org/10.1038/s41431-021-00947-1
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-021-00947-1
This article is cited by
-
Structural basis of LRPPRC–SLIRP-dependent translation by the mitoribosome
Nature Structural & Molecular Biology (2024)
-
Genomics elucidates both common and rare disease aetiology
European Journal of Human Genetics (2021)