Abstract
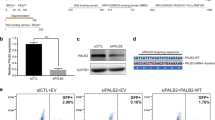
Despite routine analysis of a large panel of genes, pathogenic variants are only detected in approximately 20% of families with hereditary breast and/or ovarian cancer. Mobile element insertions (MEI) are known to cause genetic diseases in humans, but remain challenging to detect. Retrospective analysis of targeted next-generation sequencing (NGS) data from 359 patients was performed using a dedicated MEI detection pipeline. We detected one MEI in exon 9 of the PALB2 gene in a woman with a family history of breast cancer. The pathogenic variant, c.2872_2888delins114AluL2, disrupts the PALB2 coding sequence and leads to the production of a truncated protein, p.(Gln958Valfs*38). This is the first report of a pathogenic MEI in PALB2. This study illustrates that MEI analysis may help to improve molecular diagnostic yield and can be performed from targeted NGS data used for routine diagnosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
All additional data not provided in this article are available from the corresponding authors upon reasonable request. If you are interested in using the MEI detection pipeline, please contact pierre.delagrange@genodiag.com.
References
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71.
Wooster R, Bignell G, Lancaster J, Swift S, Seal S, Mangion J, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92.
Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts JK, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med. 2014;371:497–506.
Meindl A, Hellebrand H, Wiek C, Erven V, Wappenschmidt B, Niederacher D, et al. Germline mutations in breast and ovarian cancer pedigrees establish RAD51C as a human cancer susceptibility gene. Nat Genet. 2010;42:410–14.
Loveday C, Turnbull C, Ramsay E, Hughes D, Ruark E, Frankum JR, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat Genet. 2011;43:879–82.
Moretta J, Berthet P, Bonadona V, Caron O, Cohen-Haguenauer O, Colas C. et al. The French Genetic and Cancer Consortium guidelines for multigene panel analysis in hereditary breast and ovarian cancer predisposition. Bull Cancer. 2018;105:907–17.
Kazazian HH, Moran JV. Mobile DNA in health and disease. N Engl J Med. 2017;377:361–70.
Machado PM, Brandão RD, Cavaco BM, Eugénio J, Bento S, Nave M, et al. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol. 2007;25:2027–34.
Hancks DC, Kazazian HH. Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9.
Baert-Desurmont S, Coutant S, Charbonnier F, Macquere P, Lecoquierre F, Schwartz M, et al. Optimization of the diagnosis of inherited colorectal cancer using NGS and capture of exonic and intronic sequences of panel genes. Eur J Hum Genet. 2018;11:1597–602.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med J Am Coll Med Genet. 2015;17:405–24.
Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–76.
Watson CM, Camm N, Crinnion LA, Antanaviciute A, Adlard J, Markham AF, et al. Characterization and genomic localization of a SMAD4 processed pseudogene. J Mol Diagn. 2017;6:933–40.
Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–29.
Sy SM-H, Huen MSY, Zhu Y, Chen J. PALB2 regulates recombinational repair through chromatin association and oligomerization. J Biol Chem. 2009;284:18302–10.
Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–61.
Rahman N, Seal S, Thompson D, Kelly P, Renwick A, Elliott A, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–7.
Richardson ME, Chong H, Mu W, Conner BR, Hsuan V, Willett S, et al. DNA breakpoint assay reveals a majority of gross duplications occur in tandem reducing VUS classifications in breast cancer predisposition genes. Genet Med J Am Coll Med Genet. 2019;21:683–93.
Yang C, Arnold AG, Trottier M, Sonoda Y, Abu-Rustum NR, Zivanovic O, et al. Characterization of a novel germline PALB2 duplication in a hereditary breast and ovarian cancer family. Breast Cancer Res Treat. 2016;160:447–56.
Acknowledgements
We would like to thank Dr. Stéphanie Baert-Desurmont for providing the MSH2 Alu insertion fastq files.
Author contributions
FC supervised the study and contributed to analyzing data, interpreting result, and writing the manuscript. ME designed the experiments and contributed to analyzing data, interpreting result, and writing the manuscript. OA, PDLG conceived and realized bioinformatic analyses and contributed to writing the manuscript. GL performed technical experiments. NB, EG, AP contributed to analyzing data and interpreting results. CD, OC-H collected and analyzed clinical data. All authors reviewed the manuscript.
Funding
No specific funding support this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
All patients gave their informed consent for genetic research, which was approved by the “Comité de Protection des Personnes Ile de France-VI”, decision ID RCB2007-AO1347-46.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Eyries, M., Ariste, O., Legrand, G. et al. Detection of a pathogenic Alu element insertion in PALB2 gene from targeted NGS diagnostic data. Eur J Hum Genet 30, 1187–1190 (2022). https://doi.org/10.1038/s41431-022-01064-3
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-022-01064-3
This article is cited by
-
Newborn Screening for Severe T and B Cell Lymphopenia Using TREC/KREC Detection: A Large-Scale Pilot Study of 202,908 Newborns
Journal of Clinical Immunology (2024)
-
Happy 30th birthday to the European Journal of Human Genetics!
European Journal of Human Genetics (2022)