Abstract
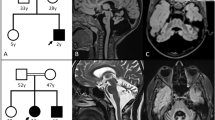
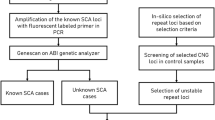
We propose an approach for the identification of mutant genes for rare diseases in single cases of unknown etiology. All genes with rare biologically significant variants sorted from individual exome data are tested further for profiling of their spatial-temporal and cell/tissue specific expression compared to that of their paralogs. We developed a simple bioinformatics tool (“Essential Paralogue by Expression” (EPbE)) for such analysis. Here, we present rare clinical forms of early ataxia with cerebellar hypoplasia. Using whole-exome sequencing and the EPbE tool, we identified two novel mutant genes previously not associated with congenital human diseases. In Family I, the unique missense mutation (p.Lys258Glu) was found in the LRCH2 gene inherited in an X-linked manner. p.Lys258Glu occurs in the evolutionarily invariant site of the leucine-rich repeat domain of LRCH2. In Family II and Family III, the identical genetic variant was found in the CSMD1 gene inherited as an autosomal-recessive trait. The variant leads to amino acid substitution p.Gly2979Ser in a highly conserved region of the complement-interacting domain of CSMD1. The LRCH2 gene for Family I patients (in which congenital cerebellar hypoplasia was associated with demyelinating polyneuropathy) is expressed in Schwann and precursor Schwann cells and predominantly over its paralogous genes in the developing cerebellar cortex. The CSMD1 gene is predominantly expressed over its paralogous genes in the cerebellum, specifically in the period of late childhood. Thus, the comparative spatial-temporal expression of the selected genes corresponds to the neurological manifestations of the disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
References
Bertini E, des Portes V, Zanni G, Santorelli F, Dionisi-Vici C, Vicari S, et al. X-linked congenital ataxia: a clinical and genetic study. Am J Med Genet. 2000;92:53–56.
Illarioshkin SN, Tanaka H, Markova ED, Nikolskaya NN, Ivanova-Smolenskaya IA, Tsuji S. X-linked nonprogressive congenital cerebellar hypoplasia: clinical description and mapping to chromosome Xq. Ann Neurol. 1996;40:75–83.
Zanni G, Calì T, Kalscheuer VM, Ottolini D, Barresi S, Lebrun N, et al. Mutation of plasma membrane Ca2+ ATPase isoform 3 in a family with X-linked congenital cerebellar ataxia impairs Ca2+ homeostasis. Proc Natl Acad Sci USA. 2012;109:14514–9.
Protasova MS, Grigorenko AP, Tyazhelova TV, Andreeva TV, Reshetov DA, Gusev FE, et al. Whole-genome sequencing identifies a novel ABCB7 gene mutation for X-linked congenital cerebellar ataxia in a large family of Mongolian ancestry. Eur J Hum Genet. 2016;24:550–5.
Tranebjaerg L, Teslovich TM, Jones M, Barmada MM, Fagerheim T, Dahl A, et al. Genome-wide homozygosity mapping localizes a gene for autosomal recessive non-progressive infantile ataxia to 20q11-q13. Hum Genet. 2003;113:293–5.
Guissart C, Li X, Leheup B, Drouot N, Montaut-Verient B, Raffo E, et al. Mutation of SLC9A1, encoding the major Na+/H+ exchanger, causes ataxia-deafness Lichtenstein-Knorr syndrome. Hum Mol Genet. 2015;24:463–70.
Duan R, Shi Y, Yu L, Zhang G, Li J, Lin Y, et al. UBA5 Mutations Cause a New Form of Autosomal Recessive Cerebellar Ataxia. PLoS One. 2016;11:e0149039.
Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF. A second fatty acid amide hydrolase with variable distribution among placental mammals. J Biol Chem. 2006;281:36569–78.
Martin HC, Gardner EJ, Samocha KE, Kaplanis J, Akawi N, Sifrim A, et al. The contribution of X-linked coding variation to severe developmental disorders. Nat Commun. 2021;12:627.
Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, et al. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J Immunol. 2006;176:4419–30.
Kraus DM, Pfenninger KH, Sanford SD, Holers VM. CSMD1 is expressed as a membrane protein on neuronal growth cones that colocalizes with F-actin and alpha-3 integrin. Mol Immunol. 2007;44:198.
Escudero-Esparza A, Kalchishkova N, Kurbasic E, Jiang WG, Blom AM. The novel complement inhibitor human CUB and Sushi multiple domains 1 (CSMD1) protein promotes factor I-mediated degradation of C4b and C3b and inhibits the membrane attack complex assembly. FASEB J. 2013;27:5083–93.
Woo JJ, Pouget JG, Zai CC, Kennedy JL. The complement system in schizophrenia: where are we now and what’s next? Mol Psychiatry. 2020;25:114–30.
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303.
McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GRS, Thormann A, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122.
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–58.
Yang J, Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43:W174–181.
Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, 1000 Genomes Project Consortium. et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43.
Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, et al. Author correction: the mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2021;590:E53.
Sim N-L, Kumar P, Hu J, Henikoff S, Schneider G, Ng PC. SIFT web server: predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012;40:W452–457.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Lindsay SJ, Xu Y, Lisgo SN, Harkin LF, Copp AJ, Gerrelli D, et al. HDBR expression: a unique resource for global and individual gene expression studies during early human brain development. Front Neuroanat. 2016;10:86.
GTEx Consortium. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45:580–5.
Miller JA, Ding S-L, Sunkin SM, Smith KA, Ng L, Szafer A, et al. Transcriptional landscape of the prenatal human brain. Nature. 2014;508:199–206.
Sirrs S, van Karnebeek CDM, Peng X, Shyr C, Tarailo-Graovac M, Mandal R, et al. Defects in fatty acid amide hydrolase 2 in a male with neurologic and psychiatric symptoms. Orphanet J Rare Dis. 2015;10:38.
Gialeli C, Gungor B, Blom AM. Novel potential inhibitors of complement system and their roles in complement regulation and beyond. Mol Immunol. 2018;102:73–83.
Kajava AV. Structural diversity of leucine-rich repeat proteins. J Mol Biol. 1998;277:519–27.
Rivière T, Bader A, Pogoda K, Walzog B, Maier-Begandt D. Structure and emerging functions of LRCH proteins in leukocyte biology. Front Cell Dev Biol. 2020;8:584134.
Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–32.
Ng ACY, Eisenberg JM, Heath RJW, Huett A, Robinson CM, Nau GJ, et al. Human leucine-rich repeat proteins: a genome-wide bioinformatic categorization and functional analysis in innate immunity. Proc Natl Acad Sci USA. 2011;108:4631–8.
Ruiz-Martínez J, Azcona LJ, Bergareche A, Martí-Massó JF, Paisán-Ruiz C. Whole-exome sequencing associates novel CSMD1 gene mutations with familial Parkinson disease. Neurol Genet. 2017;3:e177.
Mizukami T, Kohno T, Hattori M. CUB and Sushi multiple domains 3 regulates dendrite development. Neurosci Res. 2016;110:11–17.
Altenhoff AM, Train C-M, Gilbert KJ, Mediratta I, Mendes de Farias T, Moi D, et al. OMA orthology in 2021: website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Res. 2021;49:D373–D379.
Zhang X, Xu R, Zhu B, Yang X, Ding X, Duan S, et al. Syne-1 and Syne-2 play crucial roles in myonuclear anchorage and motor neuron innervation. Development. 2007;134:901–8.
Lauzier A, Bossanyi MF, Larcher R, Nassari S, Ugrankar R, Henne WM, et al. Snazarus and its human ortholog SNX25 modulate autophagic flux. J Cell Sci. 2022;135:jcs258733.
Baldwin HA, Wang C, Kanfer G, Shah HV, Velayos-Baeza A, Dulovic-Mahlow M, et al. VPS13D promotes peroxisome biogenesis. J Cell Biol. 2021;220:e202001188.
Efimova N, Korobova F, Stankewich MC, Moberly AH, Stolz DB, Wang J, et al. III Spectrin Is necessary for formation of the constricted neck of dendritic spines and regulation of synaptic activity in neurons. J Neurosci. 2017;37:6442–59.
Fujishima K, Kurisu J, Yamada M, Kengaku M. βIII spectrin controls the planarity of Purkinje cell dendrites by modulating perpendicular axon-dendrite interactions. Development. 2020;147:dev194530.
Cravatt BF, Giang DK, Mayfield SP, Boger DL, Lerner RA, Gilula NB. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87.
Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Assignment of endogenous substrates to enzymes by global metabolite profiling. Biochemistry. 2004;43:14332–9.
Müller PM, Rademacher J, Bagshaw RD, Wortmann C, Barth C, van Unen J, et al. Systems analysis of RhoGEF and RhoGAP regulatory proteins reveals spatially organized RAC1 signalling from integrin adhesions. Nat Cell Biol. 2020;22:498–511.
Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–76.
Cox BJ, Vollmer M, Tamplin O, Lu M, Biechele S, Gertsenstein M, et al. Phenotypic annotation of the mouse X chromosome. Genome Res. 2010;20:1154–64.
Zhang Y, Sloan SA, Clarke LE, Caneda C, Plaza CA, Blumenthal PD, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53.
Zhang Y, Chen K, Sloan SA, Bennett ML, Scholze AR, O’Keeffe S, et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J Neurosci. 2014;34:11929–47.
Saunders A, Macosko EZ, Wysoker A, Goldman M, Krienen FM, de Rivera H, et al. Molecular diversity and specializations among the cells of the adult mouse brain. Cell. 2018;174:1015–1030.e16.
Sapio MR, Goswami SC, Gross JR, Mannes AJ, Iadarola MJ. Transcriptomic analyses of genes and tissues in inherited sensory neuropathies. Exp Neurol. 2016;283:375–95.
Kim H-S, Lee J, Lee DY, Kim Y-D, Kim JY, Lim HJ, et al. Schwann cell precursors from human pluripotent stem cells as a potential therapeutic target for myelin repair. Stem Cell Rep. 2017;8:1714–26.
Gutierrez MA, Dwyer BE, Franco SJ. Csmd2 is a synaptic transmembrane protein that interacts with PSD-95 and is required for neuronal maturation. eNeuro 2019; 6. https://doi.org/10.1523/ENEURO.0434-18.2019.
Steen VM, Nepal C, Ersland KM, Holdhus R, Nævdal M, Ratvik SM, et al. Neuropsychological deficits in mice depleted of the schizophrenia susceptibility gene CSMD1. PLoS One. 2013;8:e79501.
Shamseldin HE, AlAbdi L, Maddirevula S, et al. Lethal variants in humans: lessons learned from a large molecular autopsy cohort. Genome Med. 2021;13:161.
Lim ET, Raychaudhuri S, Sanders SJ, Stevens C, Sabo A, MacArthur DG, et al. Rare complete knockouts in humans: population distribution and significant role in autism spectrum disorders. Neuron. 2013;77:235–42.
Funding
The genetic analysis was supported by Russian Science Foundation grant No. 19-75-30039 (MSP). The software development was supported by the Ministry of Science and Higher Education of the Russian Federation (Grant No. 075-15-2020-801) (FEG).
Author information
Authors and Affiliations
Contributions
MSP was responsible for genetic analysis, interpreting results, writing manuscript. FEG was responsible for developed EPbE software, statistical analysis and expression analysis representation. TVA was responsible for review manuscript. SAK and SNI were responsible for clinical analysis and review manuscript. EIR was responsible for supervision genetic analysis and review manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Written informed consent was obtained from all participants. The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of the Research Center of Neurology, Moscow, RF (approval number 6-2/19 of 07/10/2019).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Protasova, M.S., Gusev, F.E., Andreeva, T.V. et al. Novel genes bearing mutations in rare cases of early-onset ataxia with cerebellar hypoplasia. Eur J Hum Genet 30, 703–711 (2022). https://doi.org/10.1038/s41431-022-01088-9
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-022-01088-9
This article is cited by
-
Biallelic variants in CSMD1 are implicated in a neurodevelopmental disorder with intellectual disability and variable cortical malformations
Cell Death & Disease (2024)
-
NGS study in a sicilian case series with a genetic diagnosis for Gerstmann-Sträussler-Scheinker syndrome (PRNP, p.P102L)
Molecular Biology Reports (2023)
-
What’s new in genetics in June 2022?
European Journal of Human Genetics (2022)