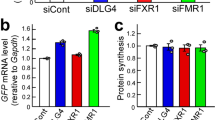
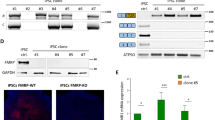
Abstract
Friedreich’s Ataxia (FRDA) is the most common hereditary ataxia and is mainly caused by biallelic GAA repeat expansion in the FXN gene. Rare patients carrying FXN point mutations or intragenic deletions are reported. We describe the first FRDA patient with a chromosome 9 segmental Uniparental isoDisomy (UPiD) unmasking a homozygous FXN expansion initially undetected by TP-PCR. The child presented with a progressive proprioceptive ataxia associated with peripheral sensory neuronopathy and severe scoliosis. Whole genome sequencing (WGS) identified a maternal segmental Uniparental Isodisomy (UPiD) encompassing FXN. Short tandem repeats analysis on WGS showed a biallelic FXN expansion. The identification of a deletion in the primer-annealing region of the TP-PCR explained the initial TP-PCR failure. This is the first documented case of FRDA caused by segmental UPiD. This case highlights the complexity of the molecular diagnosis of FRDA, and emphasises the importance of integrating results from various technical diagnostic approaches.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout

Similar content being viewed by others
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Campuzano V, Montermini L, Moltò MD, Pianese L, Cossée M, Cavalcanti F, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–7.
Cossée M, Dürr A, Schmitt M, Dahl N, Trouillas P, Allinson P, et al. Friedreich’s ataxia: point mutations and clinical presentation of compound heterozygotes. Ann Neurol. 1999;45:200–6.
Anheim M, Mariani LL, Calvas P, Cheuret E, Zagnoli F, Odent S, et al. Exonic deletions of FXN and early-onset Friedreich ataxia. Arch Neurol. 2012;69:912–6.
Bidichandani SI, Delatycki MB. Friedreich Ataxia. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Bean LJ, et al. editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993. [cited 2024 Feb 17]. Available from: http://www.ncbi.nlm.nih.gov/books/NBK1281/
Barcia G, Rachid M, Magen M, Assouline Z, Koenig M, Funalot B, et al. Pitfalls in molecular diagnosis of Friedreich ataxia. Eur J Med Genet. 2018;61:455–8.
Raman L, Dheedene A, De Smet M, Van Dorpe J, Menten B. WisecondorX: improved copy number detection for routine shallow whole-genome sequencing. Nucleic Acids Res. 2019;47:1605–14.
Dolzhenko E, Deshpande V, Schlesinger F, Krusche P, Petrovski R, Chen S, et al. ExpansionHunter: a sequence-graph-based tool to analyze variation in short tandem repeat regions. Bioinformatics. 2019;35:4754–6.
Wallace SE, Bird TD. Molecular genetic testing for hereditary ataxia. Neurol Clin Pr. 2018;8:27–32.
Dolzhenko E, van Vugt JJFA, Shaw RJ, Bekritsky MA, van Blitterswijk M, Narzisi G, et al. Detection of long repeat expansions from PCR-free whole-genome sequence data. Genome Res. 2017;27:1895–903.
Uppili B, Sharma P, Ahmad I, Sahni S, Asokachandran V, Nagaraja AB, et al. Sequencing through hyperexpanded Friedreich’s ataxia-GAA repeats by nanopore technology: implications in genotype–phenotype correlation. Brain Commun. 2023;5:fcad020.
Gana S, Valente EM. Movement disorders in genetic pediatric ataxias. Mov Disord Clin Pr. 2020;7:383–93.
Wilkinson TA, James RS, Crolla JA, Cockwell AE, Campbell PL, Temple IK. A case of maternal uniparental disomy of chromosome 9 in association with confined placental mosaicism for trisomy 9. Prenat Diagn. 1996;16:371–4.
Engel E. A new genetic concept: uniparental disomy and its potential effect, isodisomy. Am J Med Genet. 1980;6:137–43.
Tiranti V, Lamantea E, Uziel G, Zeviani M, Gasparini P, Marzella R, et al. Leigh syndrome transmitted by uniparental disomy of chromosome 9. J Med Genet. 1999;36:927–8.
Castanet M, Polak M. Spectrum of human Foxe1/TTF2 mutations. Horm Res Paediatr. 2010;73:423–9.
Papenhausen PR, Kelly CA, Harris S, Caldwell S, Schwartz S, Penton A. Clinical significance and mechanisms associated with segmental UPD. Mol Cytogenet. 2021;14:38.
del Gaudio D, Shinawi M, Astbury C, Tayeh MK, Deak KL, Raca G. Diagnostic testing for uniparental disomy: a points to consider statement from the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2020;22:1133–41.
Kotzot D. Complex and segmental uniparental disomy updated. J Med Genet. 2008;45:545–56.
Benn P. Uniparental disomy: origin, frequency, and clinical significance. Prenat Diagn. 2021;41:564–72.
Labrijn-Marks I, Somers-Bolman GM, In ’t Groen SLM, Hoogeveen-Westerveld M, Kroos MA, Ala-Mello S, et al. Segmental and total uniparental isodisomy (UPiD) as a disease mechanism in autosomal recessive lysosomal disorders: evidence from SNP arrays. Eur J Hum Genet. 2019;27:919–27.
Blanluet M, Chantot-Bastaraud S, Chambon P, Cassinari K, Vera G, Goldenberg A, et al. Recurrence of an early postzygotic rescue of an inherited unbalanced translocation resulting in mosaic segmental uniparental isodisomy of chromosome 11q in siblings. Am J Med Genet A. 2021;185:3057–61.
Scott V, Delatycki MB, Tai G, Corben LA. New and emerging drug and gene therapies for Friedreich Ataxia. CNS Drugs. 2024;38:791–805.
Acknowledgements
The authors thank the family for their participation in this study.
Funding
This manuscript did not receive any additional fund.
Author information
Authors and Affiliations
Contributions
Conceptualization: BSB, GB; Data curation: BSB, GB; Data analysis: BSB, MM, ND, JC, JMSA, MLM, ZA, JS, GB, Clinical data: CG, MR, CB, ID, GB; Writing original draft: BSB, GB; All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval
We confirm that we have read the Journal’s position on issues involved in ethical publication and affirm that this report is consistent with those guidelines.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sperelakis-Beedham, B., Gitiaux, C., Rajaoba, M. et al. Uniparental IsoDisomy: a case study on a new mechanism of Friedreich ataxia. Eur J Hum Genet 33, 137–140 (2025). https://doi.org/10.1038/s41431-024-01728-2
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41431-024-01728-2