Abstract
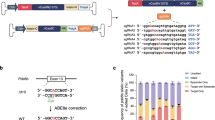
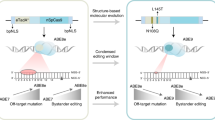
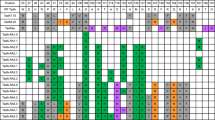
Retinitis pigmentosa (RP) associated with mutations in the rhodopsin gene (RHO) is a significant cause of blindness. Here we report on the application of adenine base editing of the c.1030C>T (p.Q344X) RHO mutation linked to RP. Using a fluorescence reporter cell system, we optimized editing by exploring base editors, sgRNA, and delivery methods. Flow cytometry, western blotting, and immunofluorescence microscopy confirmed the restoration of full-length rhodopsin after editing. DNA sequencing verified editing at the target nucleotide and the absence of bystander edits within the editing window. Polyethylenimine cationic polymer transfection of cells with a plasmid containing the NG-ABE8e adenine base editor and A6 guide RNA that placed the targeted adenine in position 6 of the editing window resulted in 31.0% gDNA sequence correction and 26.3% rhodopsin protein correction as determined by flow cytometry. Purified NG-ABE8e protein complexed with A6-sgRNA showed 32.2% gDNA editing and 44.5% rhodopsin correction. Plasmid NG-ABE8e and A6-sgRNA co-encapsulated into lipid nanoparticles (LNPs) and transfected into the reporter cell system resulted in the highest editing (42.6% gDNA editing and 65.9% rhodopsin correction). These results demonstrate the successful correction of the c.1030C>T RHO mutation and provide the foundation for base editing as a treatment for RP.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 6 print issues and online access
$259.00 per year
only $43.17 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
All the data produced in this study have been either included in the published paper or are accessible through the lead contact upon request.
References
Hanany M, Rivolta C, Sharon D. Worldwide carrier frequency and genetic prevalence of autosomal recessive inherited retinal diseases. Proc Natl Acad Sci USA. 2020;117:2710–6.
Hauswirth WW, Aleman TS, Kaushal S, Cideciyan AV, Schwartz SB, Wang L, et al. Treatment of leber congenital amaurosis due to RPE65 mutations by ocular subretinal injection of adeno-associated virus gene vector: short-term results of a phase I trial. Hum Gene Ther. 2008;19:979–90.
Maguire AM, Simonelli F, Pierce EA, Pugh EN Jr., Mingozzi F, Bennicelli J, et al. Safety and efficacy of gene transfer for Leber’s congenital amaurosis. N Engl J Med. 2008;358:2240–8.
Bainbridge JW, Smith AJ, Barker SS, Robbie S, Henderson R, Balaggan K, et al. Effect of gene therapy on visual function in Leber’s congenital amaurosis. N Engl J Med. 2008;358:2231–9.
Huang TP, Newby GA, Liu DR. Precision genome editing using cytosine and adenine base editors in mammalian cells. Nat Protoc. 2021;16:1089–128.
Choi EH, Suh S, Foik AT, Leinonen H, Newby GA, Gao XD, et al. In vivo base editing rescues cone photoreceptors in a mouse model of early-onset inherited retinal degeneration. Nat Commun. 2022;13:1830.
Muller A, Sullivan J, Schwarzer W, Wang M, Park-Windhol C, Hasler PW, et al. High-efficiency base editing in the retina in primates and human tissues. Nat Med. 2025;31:490–501.
Su J, She K, Song L, Jin X, Li R, Zhao Q, et al. In vivo base editing rescues photoreceptors in a mouse model of retinitis pigmentosa. Mol Ther Nucleic Acids. 2023;31:596–609.
Peynshaert K, Devoldere J, De Smedt SC, Remaut K. In vitro and ex vivo models to study drug delivery barriers in the posterior segment of the eye. Adv Drug Deliv Rev. 2018;126:44–57.
Chambers CZ, Soo GL, Engel AL, Glass IA, Birth Defects Research L, Frassetto A, et al. Lipid nanoparticle-mediated delivery of mRNA into the mouse and human retina and other ocular tissues. Transl Vis Sci Technol. 2024;13:7.
Gautam M, Jozic A, Su GL, Herrera-Barrera M, Curtis A, Arrizabalaga S, et al. Lipid nanoparticles with PEG-variant surface modifications mediate genome editing in the mouse retina. Nat Commun. 2023;14:6468.
Herrera-Barrera M, Ryals RC, Gautam M, Jozic A, Landry M, Korzun T, et al. Peptide-guided lipid nanoparticles deliver mRNA to the neural retina of rodents and nonhuman primates. Sci Adv. 2023;9:eadd4623.
Eygeris Y, Henderson MI, Curtis AG, Jozic A, Stoddard J, Reynaga R, et al. Preformed vesicle approach to LNP manufacturing enhances retinal mRNA delivery. Small. 2024;20:e2400815.
Hofmann KP, Lamb TD. Rhodopsin, light-sensor of vision. Prog Retin Eye Res. 2023;93:101116.
Molday RS, Moritz OL. Photoreceptors at a glance. J Cell Sci. 2015;128:4039–45.
Sung CH, Makino C, Baylor D, Nathans J. A rhodopsin gene mutation responsible for autosomal dominant retinitis pigmentosa results in a protein that is defective in localization to the photoreceptor outer segment. J Neurosci. 1994;14:5818–33.
Concepcion F, Chen J. Q344ter mutation causes mislocalization of rhodopsin molecules that are catalytically active: a mouse model of Q344ter-induced retinal degeneration. PLoS One. 2010;5:e10904.
Takita S, Jahan S, Imanishi S, Harikrishnan H, LePage D, Mann RJ, et al. Rhodopsin mislocalization drives ciliary dysregulation in a novel autosomal dominant retinitis pigmentosa knock-in mouse model. FASEB J. 2024;38:e23606.
Du SW, Newby GA, Salom D, Gao F, Menezes CR, Suh S, et al. In vivo photoreceptor base editing ameliorates rhodopsin-E150K autosomal-recessive retinitis pigmentosa in mice. Proc Natl Acad Sci USA. 2024;121:e2416827121.
Kaukonen M, McClements ME, MacLaren RE. CRISPR DNA base editing strategies for treating retinitis pigmentosa caused by mutations in rhodopsin. Genes. 2022;13:1327.
Choi H, Andersen JP, Molday RS. Expression and functional characterization of missense mutations in ATP8A2 linked to severe neurological disorders. Hum Mutat. 2019;40:2353–64.
Garces F, Jiang K, Molday LL, Stohr H, Weber BH, Lyons CJ, et al. Correlating the expression and functional activity of ABCA4 disease variants with the phenotype of patients with Stargardt disease. Invest Ophthalmol Vis Sci. 2018;59:2305–15.
Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol. 2019;37:224–6.
Kulkarni JA, Witzigmann D, Leung J, van der Meel R, Zaifman J, Darjuan MM, et al. Fusion-dependent formation of lipid nanoparticles containing macromolecular payloads. Nanoscale. 2019;11:9023–31.
Cheng MHY, Leung J, Zhang Y, Strong C, Basha G, Momeni A, et al. Induction of Bleb structures in lipid nanoparticle formulations of mRNA leads to improved transfection potency. Adv Mater. 2023;35:e2303370.
Mayuranathan T, Newby GA, Feng R, Yao Y, Mayberry KD, Lazzarotto CR, et al. Potent and uniform fetal hemoglobin induction via base editing. Nat Genet. 2023;55:1210–20.
Molday RS. Monoclonal antibodies to rhodopsin and other proteins of rod outer segments. Prog Retinal Res. 1988;8:173–209.
Doi T, Molday RS, Khorana HG. Role of the intradiscal domain in rhodopsin assembly and function. Proc Natl Acad Sci USA. 1990;87:4991–5.
Lv H, Zhang S, Wang B, Cui S, Yan J. Toxicity of cationic lipids and cationic polymers in gene delivery. J Control Release. 2006;114:100–9.
Jain S, Kumar S, Agrawal AK, Thanki K, Banerjee UC. Enhanced transfection efficiency and reduced cytotoxicity of novel lipid-polymer hybrid nanoplexes. Mol Pharm. 2013;10:2416–25.
Cullis PR, Felgner PL. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat Rev Drug Discov. 2024;23:709–22.
Akinc A, Maier MA, Manoharan M, Fitzgerald K, Jayaraman M, Barros S, et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat Nanotechnol. 2019;14:1084–7.
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63.
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–62.
Chen L, Zhang S, Xue N, Hong M, Zhang X, Zhang D, et al. Engineering a precise adenine base editor with minimal bystander editing. Nat Chem Biol. 2023;19:101–10.
Tu T, Song Z, Liu X, Wang S, He X, Xi H, et al. A precise and efficient adenine base editor. Mol Ther. 2022;30:2933–41.
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–71.
Lin Y, Wagner E, Lachelt U. Non-viral delivery of the CRISPR/Cas system: DNA versus RNA versus RNP. Biomater Sci. 2022;10:1166–92.
Kafetzis KN, Papalamprou N, McNulty E, Thong KX, Sato Y, Mironov A, et al. The effect of cryoprotectants and storage conditions on the transfection efficiency, stability, and safety of lipid-based nanoparticles for mRNA and DNA delivery. Adv Health Mater. 2023;12:e2203022.
Quagliarini E, Wang J, Renzi S, Cui L, Digiacomo L, Ferri G, et al. Mechanistic insights into the superior DNA delivery efficiency of multicomponent lipid nanoparticles: an in vitro and in vivo study. ACS Appl Mater Interfaces. 2022;14:56666–77.
Kunath K, von Harpe A, Fischer D, Petersen H, Bickel U, Voigt K, et al. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Control Release. 2003;89:113–25.
Semple SC, Akinc A, Chen J, Sandhu AP, Mui BL, Cho CK, et al. Rational design of cationic lipids for siRNA delivery. Nat Biotechnol. 2010;28:172–6.
Wen Y, Pan S, Luo X, Zhang X, Zhang W, Feng M. A biodegradable low molecular weight polyethylenimine derivative as low toxicity and efficient gene vector. Bioconjug Chem. 2009;20:322–32.
Ono R, Yasuhiko Y, Aisaki KI, Kitajima S, Kanno J, Hirabayashi Y. Exosome-mediated horizontal gene transfer occurs in double-strand break repair during genome editing. Commun Biol. 2019;2:57.
Holubowicz R, Du SW, Felgner J, Smidak R, Choi EH, Palczewska G, et al. Safer and efficient base editing and prime editing via ribonucleoproteins delivered through optimized lipid-nanoparticle formulations. Nat Biomed Eng. 2025;9:57–78.
Im SH, Jang M, Park JH, Chung HJ. Finely tuned ionizable lipid nanoparticles for CRISPR/Cas9 ribonucleoprotein delivery and gene editing. J Nanobiotechnol. 2024;22:175.
Walther J, Porenta D, Wilbie D, Seinen C, Benne N, Yang Q, et al. Comparative analysis of lipid Nanoparticle-Mediated delivery of CRISPR-Cas9 RNP versus mRNA/sgRNA for gene editing in vitro and in vivo. Eur J Pharm Biopharm. 2024;196:114207.
Pulman J, Botto C, Malki H, Ren D, Oudin P, De Cian A, et al. Direct delivery of Cas9 or base editor protein and guide RNA complex enables genome editing in the retina. Mol Ther Nucleic Acids. 2024;35:102349.
Ghoraba HH, Akhavanrezayat A, Karaca I, Yavari N, Lajevardi S, Hwang J, et al. Ocular gene therapy: a literature review with special focus on immune and inflammatory responses. Clin Ophthalmol. 2022;16:1753–71.
Athanasiou D, Aguila M, Bellingham J, Li W, McCulley C, Reeves PJ, et al. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog Retin Eye Res. 2018;62:1–23.
Acknowledgements
We thank Dr. Jonathen Yen for the NG-ABE8e ribonucleoprotein expression plasmid and Dr. Orson Moritz for the murine rhodopsin cDNA. Ryan Zhu assisted in RNP purification. Cryo-TEM data was collected at the High-Resolution Macromolecular Electron Microscopy (HRMEM) facility at UBC supported by the Canadian Foundation of Innovation and the British Columbia Knowledge Development Fund.
Funding
This study was supported by the Canadian Institutes of Health Research (CIHR) grant 175118 and an unrestricted UBC research grant to RSM and CIHR grant 148469 to PRC. MHYC was supported by the NanoMedicines Innovation Network postdoctoral fellowship in gene therapy and the CIHR Research Excellence, Diversity, and Independence (REDI) Early Career Transition Award. Y.Z. (FBD 193487) and JL was supported by a Frederick Banting and Charles Best Canada Graduate Scholarships Doctoral Award. VACP, YZ, and TM were awarded NanoMedicines Innovation Network (NMIN) graduate awards.
Author information
Authors and Affiliations
Contributions
RSM and VACP conceived the overall project. VACP, MHYC, YZ, TC, TM, JL designed the experiments, acquired the data and along with RSM, CJDR, and PRC analyzed and interpreted the data. RSM, CJDR and PRC supervised the study and provided resources. VACP and RSM wrote the original draft of the manuscript. All authors reviewed, edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
PRC has financial interests in Acuitas Therapeutics, Mesentech, and NanoVation Therapeutics. The other authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Palmgren, V.A.C., Cheng, M.H.Y., Zhang, Y. et al. Lipid nanoparticle mediated base editing of the Q344X rhodopsin mutation associated with retinitis pigmentosa. Gene Ther (2025). https://doi.org/10.1038/s41434-025-00584-z
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41434-025-00584-z