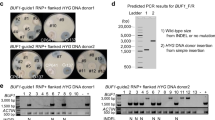
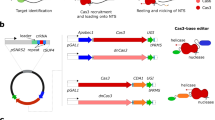
Abstract
The effective use of CRISPR technologies in emerging model organisms faces significant challenges in efficiently generating heritable mutations and in understanding the genomic consequences of induced DNA damages and the inheritance patterns of induced mutations. This study addresses these issues by 1) developing an efficient microinjection delivery method for gene editing in the microcrustacean Daphnia pulex; 2) assessing the editing dynamics of Cas9 and Cas12a nucleases in the scarlet knock-out mutants; and 3) investigating the transcriptomes of scarlet mutants to understand the pleiotropic effects of scarlet gene. Our reengineered microinjection method results in efficient biallelic editing with both nucleases. Our data suggest site-specific DNA cleavage mostly occurs in a stepwise fashion. Indels dominate the induced mutations. A few, unexpected on-site large deletions (>1 kb) are also observed. Notably, genome-wide analyses reveal no off-target mutations. Knock-in of a stop codon cassette to the scarlet locus was successful, despite complex induced mutations surrounding the target site. Moreover, extensive germline mosaicism exists in some mutants, which unexpectedly produce different phenotypes/genotypes in their asexual progeny. Lastly, our transcriptomic analyses unveil significant gene expression changes associated with scarlet knock-out and altered swimming behavior in mutants, including several genes involved in human neurodegenerative diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
DNA and RNA sequence data are available at NCBI Sequence Read Archive under project ID PRJNA1055485 and PRJNA1060702, respectively.
References
Adikusuma F, Piltz S, Corbett MA, Turvey M, McColl SR, Helbig KJ, Beard MR, Hughes J, Pomerantz RT, Thomas PQ (2018) Large deletions induced by Cas9 cleavage. Nature 560:E8–E9
Aird EJ, Lovendahl KN, St Martin A, Harris RS, Gordon WR (2018) Increasing Cas9-mediated homology-directed repair efficiency through covalent tethering of DNA repair template. Commun Biol 1:54
Akcimen F, Lopez ER, Landers JE, Nath A, Chio A, Chia R, Traynor BJ (2023) Amyotrophic lateral sclerosis: translating genetic discoveries into therapies. Nat Rev Genet 24:642–658
Alarcon M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH et al. (2008) Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet 82:150–159
Alexa A, Rahnenfuhrer J (2019) topGO: Enrichment Analysis for Gene Ontology. R Package
Altshuler I, Demiri B, Xu S, Constantin A, Yan ND, Cristescu ME (2011) An integrated multi-disciplinary approach for studying multiple stressors in freshwater ecosystems: Daphnia as a model organism. Integr Comp Biol 51:623–633
Anaka M, MacDonald CD, Barkova E, Simon K, Rostom R, Godoy RA, Haigh AJ, Meinertzhagen IA, Lloyd V (2008) The white Gene of Drosophila melanogaster Encodes a Protein with a Role in Courtship Behavior. J Neurogenet 22:243–276
Aryal NK, Wasylishen AR, Lozano G (2018) CRISPR/Cas9 can mediate high-efficiency off-target mutations in mice in vivo. Cell Death Dis 9:1099
Au V, Li-Leger E, Raymant G, Flibotte S, Chen G, Martin K, Fernando L, Doell C, Rosell FI, Wang S et al. (2019) CRISPR/Cas9 Methodology for the Generation of Knockout Deletions in Caenorhabditis elegans. G3 Bethesda 9:135–144
Auer TO, Duroure K, De Cian A, Concordet JP, Del Bene F (2014) Highly efficient CRISPR/Cas9-mediated knock-in in zebrafish by homology-independent DNA repair. Genome Res 24:142–153
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Borycz J, Borycz JA, Kubów A, Lloyd V, Meinertzhagen IA (2008) Drosophila ABC transporter mutants white, brown and scarlet have altered contents and distribution of biogenic amines in the brain. J Exp Biol 211:3454–3466
Bruce HS, Patel NH (2022) The carapace and other novel structures evolved via the cryptic persistence of serial homologs. Curr Biol 32:3792–3799
Carballar-Lejarazú R, Tushar T, Pham TB, James AA (2021) Microinjection method for Anopheles gambiae embryos. JoVE J Vis Exp 7:e62591
Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Kallberg M, Cox AJ, Kruglyak S, Saunders CT (2016) Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32:1220–1222
Colbourne JK, Hebert PDN (1996) The systematics of North American Daphnia (Crustacea: Anomopoda): A molecular phylogenetic approach. Philos Trans R Soc Lond B Biol Sci 351:349–360
Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK et al. (2011) The ecoresponsive genome of Daphnia pulex. Science 331:555–561
Cong L, Ran FA, Cox D, Lin SL, Barretto R, Habib N, Hsu PD, Wu XB, Jiang WY, Marraffini LA et al. (2013) Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339:819–823
Crawford K, Diaz Quiroz JF, Koenig KM, Ahuja N, Albertin CB, Rosenthal JJC (2020) Highly Efficient Knockout of a Squid Pigmentation Gene. Curr Biol 30:3484–3490 e3484
Curcio M, Bradke F (2018) Axon Regeneration in the Central Nervous System: Facing the Challenges from the Inside. Annu Rev Cell Dev Biol 34:495–521
Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM et al. (2021) Twelve years of SAMtools and BCFtools. Gigascience 10:giab008
Davies B (2019) The technical risks of human gene editing. Hum Reprod 34:2104–2111
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29:15–21
Ebert D (2022) Daphnia as a versatile model system in ecology and evolution. Evodevo 13:16
Evans JM, Day JP, Cabrero P, Dow JAT, Davies SA (2008) A new role for a classical gene: white transports cyclic GMP. J Exp Biol 211:890–899
Evans T (2006) Transformation and microinjection. In Transformation and microinjection, https://doi.org/10.1895/wormbook.1.108.1. The C. elegans Research Community.
Ewart GD, Cannell D, Cox GB, Howells AJ (1994) Mutational analysis of the traffic ATPase (ABC) transporters involved in uptake of eye pigment precursors in Drosophila melanogaster. Implications for structure-function relationships. J Biol Chem 269:10370–10377
Flynn JM, Chain FJJ, Schoen DJ, Cristescu ME (2017) Spontaneous mutation accumulation in Daphnia pulex in selection-free vs. competitive environments. Mol Biol Evol 34:160–173
Fu YF, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD (2013) High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat Biotechnol 31:822–826
Harris KDM, Bartlett NJ, Lloyd VK (2012) Daphnia as an Emerging Epigenetic Model Organism. Genet Res Int 2012:147892
Hefferin ML, Tomkinson AE (2005) Mechanism of DNA double-strand break repair by non-homologous end joining. DNA Repair 4:639–648
Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB et al. (2015) Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33:985–989
Hiruta C, Kakui K, Tollefsen KE, Iguchi T (2018) Targeted gene disruption by use of CRISPR/Cas9 ribonucleoprotein complexes in the water flea Daphnia pulex. Genes Cells 23:494–502
Hiruta C, Nishida C, Tochinai S (2010) Abortive meiosis in the oogenesis of parthenogenetic Daphnia pulex. Chromosome Res 18:833–840
Hiruta C, Toyota K, Miyakawa H, Ogino Y, Miyagawa S, Tatarazako N, Shaw JR, Iguchi T (2013) Development of a microinjection system for RNA interference in the water flea Daphnia pulex. BMC Biotechnol 13:96
Höijer I, Emmanouilidou A, Östlund R, van Schendel R, Bozorgpana S, Tijsterman M, Feuk L, Gyllensten U, den Hoed M, Ameur A (2022) CRISPR-Cas9 induces large structural variants at on-target and off-target sites in vivo that segregate across generations. Nat Commun 13:627
Huang S, Huang X (2023) A massively parallel approach for assessing CRISPR off-targets in vitro. Cell Rep. Methods 3:100561
Huynh TV, Hall AS, Xu S (2023) The Transcriptomic Signature of Cyclical Parthenogenesis. Genome Biol Evol 15:evad122.
Ismail NIB, Kato Y, Matsuura T, Gómez-Canela C, Barata C, Watanabe H (2021) Reduction of histamine and enhanced spinning behavior of Daphnia magna caused by scarlet mutant. Genesis 59:e23403
Ismail NIB, Kato Y, Matsuura T, Watanabe H (2018) Generation of white-eyed Daphnia magna mutants lacking scarlet function. PLoS One 13:e0205609
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337:816–821
Kato Y, Matsuura T, Watanabe H (2012) Genomic Integration and Germline Transmission of Plasmid Injected into Crustacean Daphnia magna Eggs. Plos One 7:e45318.
Kato Y, Perez CAG, Ishak NSM, Nong QD, Sudo Y, Matsuura T, Wada T, Watanabe H (2018) A 5 ‘ UTR-Overlapping LncRNA Activates the Male-Determining Gene doublesex1 in the Crustacean Daphnia magna. Curr Biol 28:1811–1817
Kato Y, Shiga Y, Kobayashi K, Tokishita S, Yamagata H, Iguchi T, Watanabe H (2011) Development of an RNA interference method in the cladoceran crustacean Daphnia magna. Dev Genes Evol 220:337–345
Keith N, Tucker AE, Jackson CE, Sung W, Lucas Lledo JI, Schrider DR, Schaack S, Dudycha JL, Ackerman M, Younge AJ et al. (2016) High mutational rates of large-scale duplication and deletion in Daphnia pulex. Genome Res 26:60–69
Kilham SS, Kreeger DA, Lynn SG, Goulden CE, Herrera L (1998) COMBO: a defined freshwater culture medium for algae and zooplankton. Hydrobiologia 377:147–159
Kim S, Kim D, Cho SW, Kim J, Kim JS (2014) Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res 24:1012–1019
Koenig MK, Hodgeman R, Riviello JJ, Chung W, Bain J, Chiriboga CA, Ichikawa K, Osaka H, Tsuji M, Gibson KM et al. (2017) Phenotype of GABA-transaminase deficiency. Neurology 88:1919–1924
Kumagai H, Nakanishi T, Matsuura T, Kato Y, Watanabe H (2017) CRISPR/Cas-mediated knock-in via non-homologous end-joining in the crustacean Daphnia magna. Plos One 12:e0186112
Li H, Durbin R (2010) Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Liang F, Han M, Romanienko PJ, Jasin M (1998) Homology-directed repair is a major double-strand break repair pathway in mammalian cells. Proc Natl Acad Sci USA 95:5172–5177
Liao Y, Smyth GK, Shi W (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30:923–930
Louro P, Ramos L, Robalo C, Cancelinha C, Dinis A, Veiga R, Pina R, Rebelo O, Pop A, Diogo L et al. (2016) Phenotyping GABA transaminase deficiency: a case description and literature review. J Inherit Metab Dis 39:743–747
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Lynch M, Gutenkunst R, Ackerman M, Spitze K, Ye Z, Maruki T, Jia Z (2017) Population Genomics of Daphnia pulex. Genetics 206:315–332
Mahato S, Morita S, Tucker AE, Liang X, Jackowska M, Friedrich M, Shiga Y, Zelhof AC (2014) Common transcriptional mechanisms for visual photoreceptor cell differentiation among Pancrustaceans. PLoS Genet 10:e1004484
Maresca M, Lin VG, Guo N, Yang Y (2013) Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res 23:539–546
Miner BE, De Meester L, Pfrender ME, Lampert W, Hairston NG (2012) Linking genes to communities and ecosystems: Daphnia as an ecogenomic model. Proc R Soc Biol Sci B 279:1873–1882
Moreno-Mateos MA, Fernandez JP, Rouet R, Vejnar CE, Lane MA, Mis E, Khokha MK, Doudna JA, Giraldez AJ (2017) CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat Commun 8:2024
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M (2007) KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 35:W182–W185
Nakanishi T, Kato Y, Matsuura T, Watanabe H (2016) TALEN-mediated knock-in via non-homologous end joining in the crustacean Daphnia magna. Combining Mult Hypothesis Sci Rep. 6:36252
Ojima Y (1958) A cytological study on the development and maturation of the parthenogenetic and sexual eggs of Daphnia pulex. Kwansei Gakuin Univ Ann Stud 6:123–171
Owens DDG, Caulder A, Frontera V, Harman JR, Allan AJ, Bucakci A, Greder L, Codner GF, Hublitz P, McHugh PJ et al. (2019) Microhomologies are prevalent at Cas9-induced larger deletions. Nucleic Acids Res 47:7402–7417
Parkinson MJ, Lilley DM (1997) The junction-resolving enzyme T7 endonuclease I: quaternary structure and interaction with DNA. J Mol Biol 270:169–178
Peippo M, Ignatius J (2011) Pitt-Hopkins Syndrome. Pitt-Hopkins Syndr 2:171–180
Rasys AM, Park S, Ball RE, Alcala AJ, Lauderdale JD, Menke DB (2019) CRISPR-Cas9 Gene Editing in Lizards through Microinjection of Unfertilized Oocytes. Cell Rep. 28:2288–2292.e2283
Riesenberg S, Kanis P, Macak D, Wollny D, Dusterhoft D, Kowalewski J, Helmbrecht N, Maricic T, Paabo S (2023) Efficient high-precision homology-directed repair-dependent genome editing by HDRobust. Nat Methods 20:1388–1399
Ringrose L (2009) Transgenesis in Drosophila melanogaster. Methods Mol Biol 561:3–19
Rivera AS, Pankey MS, Plachetzki DC, Villacorta C, Syme AE, Serb JM, Omilian AR, Oakley TH (2010) Gene duplication and the origins of morphological complexity in pancrustacean eyes, a genomic approach. BMC Evol Biol 10:123
Rodgers K, McVey M (2016) Error-Prone Repair of DNA Double-Strand Breaks. J Cell Physiol 231:15–24
Schimmel J, Munoz-Subirana N, Kool H, van Schendel R, van der Vlies S, Kamp JA, de Vrij FMS, Kushner SA, Smith GCM, Boulton SJ et al. (2023) Modulating mutational outcomes and improving precise gene editing at CRISPR-Cas9-induced breaks by chemical inhibition of end-joining pathways. Cell Rep. 42:112019
Sekelsky J (2017) DNA Repair in Drosophila: Mutagens, Models, and Missing Genes. Genetics 205:471–490
Sfeir A, Symington LS (2015) Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci 40:701–714
Sharma A, Pham MN, Reyes JB, Chana R, Yim WC, Heu CC, Kim D, Chaverra-Rodriguez D, Rasgon JL, Harrell RA et al. (2022) Cas9-mediated gene editing in the black-legged tick, Ixodes scapularis, by embryo injection and ReMOT Control. iScience, https://doi.org/10.1016/j.isci.2022.103781
Sharon E, Chen SA, Khosla NM, Smith JD, Pritchard JK, Fraser HB (2018) Functional Genetic Variants Revealed by Massively Parallel Precise Genome Editing. Cell 175:544–557.e516
Shaw JR, Colbourne JK, Davey JC, Glaholt SP, Hampton TH, Chen CY, Folt CL, Hamilton JW (2007) Gene response profiles for Daphnia pulex exposed to the environmental stressor cadmium reveals novel crustacean metallothioneins. BMC Genomics 8:477
Shin HY, Wang C, Lee HK, Yoo KH, Zeng X, Kuhns T, Yang CM, Mohr T, Liu C, Hennighausen L (2017) CRISPR/Cas9 targeting events cause complex deletions and insertions at 17 sites in the mouse genome. Nat Commun 8:15464
Shirai Y, Piulachs M-D, Belles X, Daimon T (2022) DIPA-CRISPR is a simple and accessible method for insect gene editing. Cell Rep Methods 2:100215
Shy BR, Vykunta VS, Ha A, Talbot A, Roth TL, Nguyen DN, Pfeifer WG, Chen YY, Blaeschke F, Shifrut E et al. (2023) High-yield genome engineering in primary cells using a hybrid ssDNA repair template and small-molecule cocktails. Nat Biotechnol 41:521–531
Snyman M, Huynh TV, Smith MT, Xu S (2021) The genome-wide rate and spectrum of EMS-induced heritable mutations in the microcrustacean Daphnia: on the prospect of forward genetics. Heredity 127:535–545
Snyman M, Xu S (2023) Transcriptomics and the origin of obligate parthenogenesis. Heredity 131:119–129
Toyota K, Miyagawa S, Ogino Y, Iguchi T (2016) Microinjection-based RNA interference method in the water flea, Daphnia pulex and Daphnia magna. In RNA Interference. IntechOpen.
Trible W, Olivos-Cisneros L, McKenzie SK, Saragosti J, Chang NC, Matthews BJ, Oxley PR, Kronauer DJC (2017) orco Mutagenesis Causes Loss of Antennal Lobe Glomeruli and Impaired Social Behavior in Ants. Cell 170:727–735.e710
Wang H, Xu S (2021) Something old, something new, a DNA preparation procedure for long-read genomic sequencing. Genome 65:53–56
Wersebe MJ, Weider LJ (2023) Resurrection genomics provides molecular and phenotypic evidence of rapid adaptation to salinization in a keystone aquatic species. Proc Natl Acad Sci USA 120:e2217276120
Xu S, Huynh TV, Snyman M (2022) The transcriptomic signature of obligate parthenogenesis. Heredity 128:132–138
Xu S, Omilian AR, Cristescu ME (2011) High rate of large-scale hemizygous deletions in asexually propagating Daphnia: omplications for the evolution of sex. Mol Biol Evol 28:335–342
Xu S, Pham TP, Neupane S (2020) Delivery methods for CRISPR/Cas9 gene editing in crustaceans. Mar Life Sci Technol 2:1–5
Xu S, Spitze K, Ackerman MS, Ye Z, Bright L, Keith N, Jackson CE, Shaw JR, Lynch M (2015) Hybridization and the origin of contagious asexuality in Daphnia pulex. Mol Biol Evol 32:3215–3225
Ye Z, Pfrender ME, Lynch M (2023) Evolutionary Genomics of Sister Species Differing in Effective Population Sizes and Recombination Rates. Genome Biol Evol 15:evad202
Ye Z, Xu S, Spitze K, Asselman J, Jiang X, Ackerman MS, Lopez J, Harker B, Raborn RT, Thomas WK et al. (2017) A new reference genome assembly for the microcrustacean Daphnia pulex. G3 Bethesda 7:1405–1416
Zaffagnini F, Sabelli B (1972) Karyologic observations on the maturation of the summer and winter eggs of Daphnia pulex and Daphnia middendorffiana. Chromosoma 36:193–203
Acknowledgements
We would like to thank J. Dittmer and P. Wright for their assistance with swimming behavior experiments. We also thank Michael Pfrender for his comments on an early draft of this manuscript. This work is supported by NIH grant R35GM133730, NSF CAREER grant MCB-2042490/2348390, NSF EDGE grant 2220695/2324639 to SX.
Author information
Authors and Affiliations
Contributions
SX designed research, performed experiments, analyzed data, and wrote the manuscript. SN, HW, and TPP performed microinjection experiments, analyzed mutant and genomic data, and contributed to the writing of the manuscript. MS performed RNA-seq experiments, analyzed RNA-seq data, and contributed to the writing of manuscript. TH analyzed knock-in mutant genomic sequences. LW performed behavior experiments of scarlet mutants and analyzed data.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Associate editor: Sara Goodacre.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, S., Neupane, S., Wang, H. et al. The mosaicism of Cas-induced mutations and pleiotropic effects of scarlet gene in an emerging model system. Heredity 134, 221–233 (2025). https://doi.org/10.1038/s41437-025-00750-4
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41437-025-00750-4