Abstract
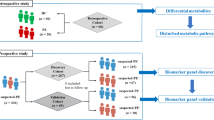
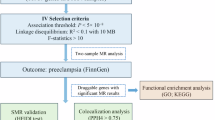
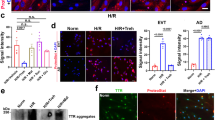
Preeclampsia (PE) is a heterogeneous disease that seriously affects the health of mothers and fetuses. Lack of detection assays, its diagnosis and intervention are often delayed when the clinical symptoms are atypical. Using personalized pathway-based analysis and machine learning algorithms, we built a PE diagnosis model consisting of nine core pathways using multiple cohorts from the Gene Expression Omnibus database. The model showed an area under the receiver operating characteristic (AUROC) curve of 0.959 with the data from the placental tissue samples in the development cohort. In the two validation cohorts, the AUROCs were 0.898 and 0.876, respectively. The model also performed well with the maternal plasma data in another validation cohort (AUROC: 0.815). Moreover, we identified tyrosine-protein kinase Lck (LCK) as the hub gene in this model and found that LCK and pLCK proteins were downregulated in placentas from PE patients. The pathway-level model for PE can provide a novel direction to develop molecular diagnostic assay and investigate potential mechanisms of PE in future studies.

This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Data availability
The gene expression data used in the present study are publicly available in the GEO database (http://www.ncbi.nlm.nih.gov/geo).
References
Chappell LC, Cluver CA, Kingdom J, Tong S. Pre-eclampsia. Lancet. 2021;398:341–54.
Duhig KE, Myers J, Seed PT, Sparkes J, Lowe J, Hunter RM, et al. Placental growth factor testing to assess women with suspected pre-eclampsia: a multicentre, pragmatic, stepped-wedge cluster-randomised controlled trial. Lancet. 2019;393:1807–18.
Fox A, McHugh S, Browne J, Kenny LC, Fitzgerald A, Khashan AS, et al. Estimating the Cost of Preeclampsia in the Healthcare System: Cross-Sectional Study Using Data From SCOPE Study (Screening for Pregnancy End Points). Hypertension. 2017;70:1243–9.
Navajas R, Corrales F, Paradela A. Quantitative proteomics-based analyses performed on pre-eclampsia samples in the 2004-2020 period: a systematic review. Clin Proteom. 2021;18:6.
Poon LC, Shennan A, Hyett JA, Kapur A, Hadar E, Divakar H, et al. The International Federation of Gynecology and Obstetrics (FIGO) initiative on pre-eclampsia: A pragmatic guide for first-trimester screening and prevention. Int J Gynaecol Obstet. 2019;145:1–33.
Gupta S, Arango-Argoty G, Zhang L, Pruden A, Vikesland P. Identification of discriminatory antibiotic resistance genes among environmental resistomes using extremely randomized tree algorithm. Microbiome. 2019;7:123.
Clarke R, Ressom HW, Wang A, Xuan J, Liu MC, Gehan EA, et al. The properties of high-dimensional data spaces: implications for exploring gene and protein expression data. Nat Rev Cancer. 2008;8:37–49.
Wilms M, Handels H, Ehrhardt J. Multi-resolution multi-object statistical shape models based on the locality assumption. Med Image Anal. 2017;38:17–29.
Vinga S. Structured sparsity regularization for analyzing high-dimensional omics data. Brief Bioinform. 2021;22:77–87.
Wu K, Liu J, Wang S. Reconstructing Networks from Profit Sequences in Evolutionary Games via a Multiobjective Optimization Approach with Lasso Initialization. Sci Rep. 2016;6:37771.
Park KS, Kim SH, Oh JH, Kim SY. Highly accurate diagnosis of papillary thyroid carcinomas based on personalized pathways coupled with machine learning. Brief Bioinform. 2021;22:bbaa336.
Friedman J, Hastie T, Tibshirani R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J Stat Softw. 2010;33:1–22.
Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc B. 2005;67:301–20.
Glaab E. Using prior knowledge from cellular pathways and molecular networks for diagnostic specimen classification. Brief Bioinform. 2016;17:440–52.
Drier Y, Sheffer M, Domany E. Pathway-based personalized analysis of cancer. Proc Natl Acad Sci USA. 2013;110:6388–93.
Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–10.
Magee LA, Brown MA, Hall DR, Gupte S, Hennessy A, Karumanchi SA, et al. The 2021 International Society for the Study of Hypertension in Pregnancy classification, diagnosis & management recommendations for international practice. Pregnancy Hypertens. 2022;27:148–69.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, et al. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–9.
Nishimura D. Biotech Software & Internet Report. Open Access Libr. 2001;2:117–20.
Gillespie M, Jassal B, Stephan R, Milacic M, Rothfels K, Senff-Ribeiro A, et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022;50:D687–D92.
Sill M, Hielscher T, Becker N, Zucknick M. c060: Extended Inference with Lasso and Elastic-Net Regularized Cox and Generalized Linear Models. J Stat Softw. 2014;62:1–22.
Hernández-Orallo J, Flach P, Ferri C. A unified view of performance metrics: translating threshold choice into expected classification loss. J Mach Learn Res. 2012;13:2813–69.
DeVries Z, Locke E, Hoda M, Moravek D, Phan K, Stratton A, et al. Using a national surgical database to predict complications following posterior lumbar surgery and comparing the area under the curve and F1-score for the assessment of prognostic capability. Spine J. 2021;21:1135–42.
Rahmatallah Y, Zybailov B, Emmert-Streib F, Glazko G. GSAR: Bioconductor package for Gene Set analysis in R. BMC Bioinforma. 2017;18:61.
Rahmatallah Y, Emmert-Streib F, Glazko G. Gene Sets Net Correlations Analysis (GSNCA): a multivariate differential coexpression test for gene sets. Bioinformatics. 2014;30:360–8.
He A, Wang J, Yang X, Liu J, Yang X, Wang G, et al. Screening of differentially expressed proteins in placentas from patients with late-onset preeclampsia. Proteom Clin Appl. 2022;16:e2100053.
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling Tumor Infiltrating Immune Cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinforma. 2013;14:7.
Leavey K, Benton SJ, Grynspan D, Bainbridge SA, Morgen EK, Cox BJ. Gene markers of normal villous maturation and their expression in placentas with maturational pathology. Placenta. 2017;58:52–9.
Guo L, Tsai SQ, Hardison NE, James AH, Motsinger-Reif AA, Thames B, et al. Differentially expressed microRNAs and affected biological pathways revealed by modulated modularity clustering (MMC) analysis of human preeclamptic and IUGR placentas. Placenta. 2013;34:599–605.
Leavey K, Wilson SL, Bainbridge SA, Robinson WP, Cox BJ. Epigenetic regulation of placental gene expression in transcriptional subtypes of preeclampsia. Clin Epigenetics. 2018;10:28.
Tsai S, Hardison NE, James AH, Motsinger-Reif AA, Bischoff SR, Thames BH, et al. Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta. 2011;32:175–82.
Textoris J, Ivorra D, Ben Amara A, Sabatier F, Menard JP, Heckenroth H, et al. Evaluation of current and new biomarkers in severe preeclampsia: a microarray approach reveals the VSIG4 gene as a potential blood biomarker. PLoS One. 2013;8:e82638.
Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27.
Zeisler H, Llurba E, Chantraine F, Vatish M, Staff AC, Sennstrom M, et al. Predictive Value of the sFlt-1:PlGF Ratio in Women with Suspected Preeclampsia. N. Engl J Med. 2016;374:13–22.
Gallo DM, Wright D, Casanova C, Campanero M, Nicolaides KH. Competing risks model in screening for preeclampsia by maternal factors and biomarkers at 19-24 weeks’ gestation. Am J Obstet Gynecol. 2016;214:619.e1–17.
Benny PA, Alakwaa FM, Schlueter RJ, Lassiter CB, Garmire LX. A review of omics approaches to study preeclampsia. Placenta. 2020;92:17–27.
Wei Z, Zhang Y, Weng W, Chen J, Cai H. Survey and comparative assessments of computational multi-omics integrative methods with multiple regulatory networks identifying distinct tumor compositions across pan-cancer data sets. Brief Bioinform. 2021;22:bbaa102.
Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10:e0118432.
Yart L, Roset Bahmanyar E, Cohen M, Martinez de Tejada B. Role of the Uteroplacental Renin-Angiotensin System in Placental Development and Function, and Its Implication in the Preeclampsia Pathogenesis. Biomedicines. 2021;9:1332.
Saei H, Govahi A, Abiri A, Eghbali M, Abiri M. Comprehensive transcriptome mining identified the gene expression signature and differentially regulated pathways of the late-onset preeclampsia. Pregnancy Hypertens. 2021;25:91–102.
Huang Y, Zheng XD, Li H. Protective role of SIRT1-mediated Sonic Hedgehog signaling pathway in the preeclampsia rat models. J Assist Reprod Genet. 2021;38:1843–51.
Miller D, Motomura K, Galaz J, Gershater M, Lee ED, Romero R, et al. Cellular immune responses in the pathophysiology of preeclampsia. J Leukoc Biol. 2022;111:237–60.
He X, Zhao L, Yue L, Zhang W, Wang W, Fu Y, et al. The relationship between IGF1 and the expression spectrum of miRNA in the placenta of preeclampsia patients. Ginekol Pol. 2019;90:596–603.
Speake PF, Glazier JD, Ayuk PT, Reade M, Sibley CP, D’Souza SWL. -Arginine transport across the basal plasma membrane of the syncytiotrophoblast of the human placenta from normal and preeclamptic pregnancies. J Clin Endocrinol Metab. 2003;88:4287–92.
Nicola C, Lala PK, Chakraborty C. Prostaglandin E2-mediated migration of human trophoblast requires RAC1 and CDC42. Biol Reprod. 2008;78:976–82.
Saso J, Shields SK, Zuo Y, Chakraborty C. Role of Rho GTPases in human trophoblast migration induced by IGFBP1. Biol Reprod. 2012;86:1–9.
Wang H, Liu ML, Chu C, Yu SJ, Li J, Shen HC, et al. Paeonol alleviates placental inflammation and apoptosis in preeclampsia by inhibiting the JAK2/STAT3 signaling pathway. Kaohsiung J Med Sci. 2022;38:1103–12.
Palacios EH, Weiss A. Function of the Src-family kinases, Lck and Fyn, in T-cell development and activation. Oncogene. 2004;23:7990–8000.
Courtney AH, Amacher JF, Kadlecek TA, Mollenauer MN, Au-Yeung BB, Kuriyan J, et al. A Phosphosite within the SH2 Domain of Lck Regulates Its Activation by CD45. Mol Cell. 2017;67:498–511.e6.
Hauck F, Randriamampita C, Martin E, Gerart S, Lambert N, Lim A, et al. Primary T-cell immunodeficiency with immunodysregulation caused by autosomal recessive LCK deficiency. J Allergy Clin Immunol. 2012;130:1144–52.e11.
Laresgoiti-Servitje E. A leading role for the immune system in the pathophysiology of preeclampsia. J Leukoc Biol. 2013;94:247–57.
Saito S, Tsuda S, Nakashima A. T cell immunity and the etiology and pathogenesis of preeclampsia. J Reprod Immunol. 2023;159:104125.
Ji ZS, Jiang H, Xie Y, Wei QP, Yin XF, Ye JH, et al. Chemerin promotes the pathogenesis of preeclampsia by activating CMKLR1/p-Akt/CEBPa axis and inducing M1 macrophage polarization. Cell Biol Toxicol. 2021. https://doi.org/10.1007/s10565-021-09636-7.
Acknowledgements
The authors thank all participants in this study, including those who donated placental tissues and those who provided sample data in the GEO database. The graphical abstract was created with BioRender (https://www.biorender.com/). In addition, the authors thank Beijing GAP BioTechnology Co., Ltd. for their assistance with the bioinformatics analysis.
Funding
This work was supported by grants from the Natural Science Foundation of Guangdong Province, China (No. 2023A1515012741), Clinical Frontier Technology Program of the First Affiliated Hospital of Jinan University, China (No. JNU1AF-CFTP-2022-a01209), and Research on the Mechanisms and Clinical Implications of Ferroptosis in Preeclampsia (No. 2023A03J1009). There was no role of the funding body in the study design, data collection and analysis, and manuscript writing.
Author information
Authors and Affiliations
Contributions
Ruiman Li, Feng Gao, Ruiling Yan, Qiao Zhang, Andong He, Daiqiang Lu, and Jia Liu designed the study. Andong He, Ka Cheuk Yip, Daiqiang Lu, Jia Liu, Zunhao Zhang, Xiufang Wang, Yifeng Liu, and Yiling Wei performed the analysis. Andong He and Ka Cheuk Yip wrote the manuscript. Ruiman Li, Feng Gao, Ruiling Yan, and Qiao Zhang assisted in revising the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, A., Yip, K.C., Lu, D. et al. Construction of a pathway-level model for preeclampsia based on gene expression data. Hypertens Res 47, 2521–2531 (2024). https://doi.org/10.1038/s41440-024-01753-0
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41440-024-01753-0