Abstract
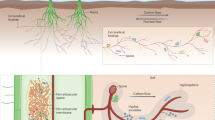
The interplay between nutrient availability and arbuscular mycorrhizal fungi (AMF) symbiosis during plant growth exhibits intricate complexity. In this study, we employ integrated physiological, transcriptomic, proteomic, and metabolomic analyses to investigate how sugarcane differentially adapts to nitrogen (N) fertilization and AMF colonization. Under nitrogen stress conditions, AMF colonization significantly enhances sugarcane growth, increasing plant height, stem diameter, and biomass while stimulating root exudation and rhizospheric nutrient mobilization-particularly available N, phosphorus (P), and potassium (K). Multi-omics analyses reveal that AMF induces nitrogen-dependent metabolic reprogramming in sugarcane roots, activating pathways such as carbohydrate and lipid metabolic pathways while suppressing butanoate and ascorbate metabolism. Weighted gene co-expression network analysis (WGCNA) identifies key root modules strongly correlated with soil N, P, and K availability, indicating AMF-mediated coordination of nutrient acquisition strategies. Field trials demonstrate that AMF boost sugarcane yield under nitrogen stress by enhancing root elongation and carbon partitioning for sucrose accumulation. Temporal integration of transcriptomic and metabolomic data highlights flavonoid biosynthesis as a persistently activated pathway across growth stages, potentially facilitating AMF symbiosis and stress resilience. Our findings elucidate how sugarcane optimizes AMF-mediated nutrient acquisition under nitrogen stress through root transcriptional and metabolic adjustments, providing insights for sustainable crop nutrient management.
Similar content being viewed by others
Data availability
The RNA sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under accession numbers PRJNA967801 (https://www.ncbi.nlm.nih.gov/bioproject/term=PRJNA967801) and Genome Sequence Archive under accession numbers PRJCA041107 (https://ngdc.cncb.ac.cn/gsub/submit/bioproject/subPRO060471/overview). The proteomics data are available through ProjectID IPX0012075001 (https://www.iprox.cn/page/PCV010.html?UploadPage=1&subprojectId=IPX0012075001) in the iProX. Source data are provided with this manuscript.
References
Coskun, D., Britto, D. T., Shi, W. & Kronzucker, H. J. Nitrogen transformations in modern agriculture and the role of biological nitrification inhibition. Nat. Plants 3, 17074 (2017).
You, L. et al. Optimized agricultural management reduces global cropland nitrogen losses to air and water. Nat. Food 5, 995–1004 (2024).
Robinson, N., Vogt, J., Lakshmanan, P. & Schmidt, S. Nitrogen physiology of sugarcane. Sugarcane: Physiology, Biochemistry, and Functional Biology, 169-195 https://doi.org/10.1002/9781118771280.ch8 (2013).
Yang, W., Li, Z., Wang, J., Wu, P. & Zhang, Y. Crop yield, nitrogen acquisition and sugarcane quality as affected by interspecific competition and nitrogen application. Field Crop Res. 146, 44–50 (2013).
Cardozo, N. P., de Oliveira Bordonal, R. & La, S. N. Sustainable intensification of sugarcane production under irrigation systems, considering climate interactions and agricultural efficiency. J. Clean. Prod. 204, 861–871 (2018).
Vandenberghe, L. P. S. et al. Beyond sugar and ethanol: the future of sugarcane biorefineries in Brazil. Renew. Sustain. Energy Rev. 167, 112721 (2022).
Parniske, M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 6, 763–775 (2008).
Wang, J. et al. Arbuscular mycorrhizal fungi regulate the diversity–invasion resistance relationship by influencing the role of complementarity and selection effects. N. Phytol. 246, 317–330 (2025).
Willing, C. E., Wan, J., Yeam, J. J., Cessna, A. M. & Peay, K. G. Arbuscular mycorrhizal fungi equalize differences in plant fitness and facilitate plant species coexistence through niche differentiation. Nat. Ecol. Evol. 8, 2058–2071 (2024).
Wang, G., Jin, Z., George, T. S., Feng, G. & Zhang, L. Arbuscular mycorrhizal fungi enhance plant phosphorus uptake through stimulating hyphosphere soil microbiome functional profiles for phosphorus turnover. N. Phytol. 238, 2578–2593 (2023).
Wang, L., Zhang, L., George, T. S. & Feng, G. Hyphosphere core taxa link plant-arbuscular mycorrhizal fungi combinations to soil organic phosphorus mineralization. Soil Biol. Biochem. 201, 109647 (2025).
Nagy, R., Drissner, D., Amrhein, N., Jakobsen, I. & Bucher, M. Mycorrhizal phosphate uptake pathway in tomato is phosphorus-repressible and transcriptionally regulated. N. Phytol. 181, 950–959 (2009).
DUAN, S. et al. The interplay of direct and mycorrhizal pathways for plants to efficiently acquire phosphorus from soil. Front. Agric. Sci. Eng. 12, 47–56 (2025).
Martin, F. M. & van der Heijden, M. G. A. The mycorrhizal symbiosis: research frontiers in genomics, ecology, and agricultural application. N. Phytol. 242, 1486–1506 (2024).
Sun, K. et al. Hyphosphere microorganisms facilitate hyphal spreading and root colonization of plant symbiotic fungus in ammonium-enriched soil. ISME J. 17, 1626–1638 (2023).
Wang, F. et al. Exploring the secrets of hyphosphere of arbuscular mycorrhizal fungi: processes and ecological functions. Plant Soil 481, 1–22 (2022).
Monika, Y. N. et al. Arbuscular Mycorrhizal fungi: a potential candidate for nitrogen fixation. in (eds Vaishnav A., Arya S. S. & Choudhary D. K.) Plant Stress Mitigators: Action and Application, 217–234 (Springer Nature, 2022).
Tian, C. et al. Regulation of the nitrogen transfer pathway in the arbuscular mycorrhizal symbiosis: gene characterization and the coordination of expression with nitrogen flux. Plant Physiol. 153, 1175–1187 (2010).
Sajjad, N. et al. 12 - Nitrogen uptake, assimilation, and mobilization in plants under abiotic stress. in (eds Roychoudhury, A., Tripathi, D. K. & Deshmukh, R.) Transporters and Plant Osmotic Stress, 215–233 (Academic Press, 2021).
Wipf, D. et al. Trading on the arbuscular mycorrhiza market: from arbuscules to common mycorrhizal networks. N. Phytol. 223, 1127–1142 (2019).
Chen, Y. et al. Transcriptomic analysis of nitrogen metabolism pathways in Klebsiella aerogenes under nitrogen-rich conditions. Front. Microbiol. 15, 1323160 (2024).
Hetrick, B. A. D. Mycorrhizas and root architecture. Experientia 47, 355–362 (1991).
Nair, A., Thulasiram, H. V. & Bhargava, S. Role of jasmonate in modulation of mycorrhizae-induced resistance against fungal pathogens. in (eds Champion, A. & Laplaze, L.) Jasmonate in Plant Biology: Methods and Protocols, 109–115 (Springer, 2020).
Lanfranco, L., Fiorilli, V. & Gutjahr, C. Partner communication and role of nutrients in the arbuscular mycorrhizal symbiosis. N. Phytol. 220, 1031–1046 (2018).
Manck-Götzenberger, J. & Requena, N. Arbuscular mycorrhiza symbiosis induces a major transcriptional reprogramming of the potato sweet sugar transporter family. Front. Plant Sci. 7, 487 (2016).
Zhou, J. et al. SYMRK significantly affected AMF symbiosis and plant growth in maize. Plant Sci. 353, 112427 (2025).
Ferrol, N., Azcón-Aguilar, C. & Pérez-Tienda, J. Review: arbuscular mycorrhizas as key players in sustainable plant phosphorus acquisition: an overview on the mechanisms involved. Plant Sci. 280, 441–447 (2019).
Wang, S. et al. OsNLP3 and OsPHR2 orchestrate direct and mycorrhizal pathways for nitrate uptake by regulating NAR2.1-NRT2s complexes in rice. Proc. Natl. Acad. Sci. USA 122, e1878622174 (2025).
Cerda, A. & Alvarez, J. M. Insights into molecular links and transcription networks integrating drought stress and nitrogen signaling. N. Phytol. 241, 560–566 (2024).
Farhan, M. et al. Plant nitrogen metabolism: balancing resilience to nutritional stress and abiotic challenges. Phyton Int. J. Exp. Bot. 93, 581–609 (2024).
Shanks, C. M. et al. Nitrogen sensing and regulatory networks: it’s about time and space. Plant Cell 36, 1482–1503 (2024).
Forde, B. G. Nitrogen signalling pathways shaping root system architecture: an update. Curr. Opin. Plant Biol. 21, 30–36 (2014).
Isah, T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 52, 39 (2019).
Johnson, N. C., Wilson, G. W. T., Bowker, M. A., Wilson, J. A. & Miller, R. M. Resource limitation is a driver of local adaptation in mycorrhizal symbioses. Proc. Natl. Acad. Sci. USA 107, 2093–2098 (2010).
Treseder, K. K. A. A meta-analysis of mycorrhizal responses to nitrogen, phosphorus, and atmospheric CO2 in field studies. New Phytol. 164, 347–355 (2004).
Govindarajulu, M. et al. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 435, 819–823 (2005).
Hodge, A. & Fitter, A. H. Substantial nitrogen acquisition by arbuscular mycorrhizal fungi from organic material has implications for N cycling. Proc. Natl. Acad. Sci. USA 107, 13754–13759 (2010).
Lin, F. et al. Emerging roles of phosphoinositide-associated membrane trafficking in plant stress responses. J. Genet Genom. 49, 726–734 (2022).
Kumar, V., Thakur, J. K. & Prasad, M. Histone acetylation dynamics regulating plant development and stress responses. Cell Mol. Life Sci. 78, 4467–4486 (2021).
Wang, Q., Yung, W., Wang, Z. & Lam, H. The histone modification H3K4me3 marks functional genes in soybean nodules. Genomics 112, 5282–5294 (2020).
Gutjahr, C. et al. Rice perception of symbiotic arbuscular mycorrhizal fungi requires the karrikin receptor complex. Science 350, 1521–1524 (2015).
Anani, O. A., Abel, I., Olomukoro, J. O. & Onyeachu, I. B. Insights to proteomics and metabolomics metal chelation in food crops. J. Proteins Proteom. 13, 159–173 (2022).
Wu, D., Saleem, M., He, T. & He, G. The mechanism of metal homeostasis in plants: a new view on the synergistic regulation pathway of membrane proteins, lipids and metal ions. Membranes 11, 984 (2021).
Donaldson, J. G. & Jackson, C. L. ARF family G proteins and their regulators: roles in membrane transport, development and disease. Nat. Rev. Mol. Cell Bio. 12, 362–375 (2011).
Gao, Y. et al. Analysis of the molecular and biochemical mechanisms involved in the symbiotic relationship between arbuscular mycorrhiza fungi and Manihot esculenta Crantz. Front. Plant Sci. 14, 1130924 (2023).
Limpens, E. & Geurts, R. Transcriptional regulation of nutrient exchange in arbuscular mycorrhizal symbiosis. Mol. Plant 11, 1421–1423 (2018).
Kabir, A. H. et al. Arbuscular mycorrhizal fungi alleviate Fe-deficiency symptoms in sunflower by increasing iron uptake and its availability along with antioxidant defense. Plant Physiol. Biochem. 150, 254–262 (2020).
Moreno Jiménez, E., Ferrol, N., Corradi, N., Peñalosa, J. M. & Rillig, M. C. The potential of arbuscular mycorrhizal fungi to enhance metallic micronutrient uptake and mitigate food contamination in agriculture: prospects and challenges. N. Phytol. 242, 1441–1447 (2024).
Rahman, M. A. et al. Arbuscular mycorrhizal symbiosis mitigates iron (Fe)-deficiency retardation in Alfalfa (Medicago sativa L.) through the enhancement of Fe accumulation and sulfur-assisted antioxidant defense. Int. J. Mol. Sci. 21, 6 (2020).
Jin, X. et al. Adaptation strategies of seedling root response to nitrogen and phosphorus addition. Plants 13, 536 (2024).
Yang, T. et al. Global transcriptomic analysis reveals candidate genes associated with different phosphorus acquisition strategies among soybean varieties. Front. Plant Sci. 13, 0–0 (2022).
Luan, M. et al. Transport and homeostasis of potassium and phosphate: limiting factors for sustainable crop production. J. Exp. Bot. 68, 3091–3105 (2017).
Wang, Y., Chen, Y. & Wu, W. Potassium and phosphorus transport and signaling in plants. J. Integr. Plant Biol. 63, 34–52 (2021).
DiTusa, S. F. et al. A member of the Phosphate transporter 1 (Pht1) family from the arsenic-hyperaccumulating fern Pteris vittata is a high-affinity arsenate transporter. N. Phytol. 209, 762–772 (2016).
Lu, Y. et al. Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis. Nat. Commun. 13, 5682 (2022).
Yang, C. et al. Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 56, 414–424 (2014).
Zhang, Y. & Fernie, A. R. The role of TCA cycle enzymes in plants. Adv. Biol. 7, 2200238 (2023).
Huang, S., Braun, H., Gawryluk, R. M. R. & Millar, A. H. Mitochondrial complex II of plants: subunit composition, assembly, and function in respiration and signaling. Plant J. 98, 405–417 (2019).
Luginbuehl, L. H. et al. Fatty acids in arbuscular mycorrhizal fungi are synthesized by the host plant. Science 356, 1175–1178 (2017).
Brinkmann-Chen, S. et al. General approach to reversing ketol-acid reductoisomerase cofactor dependence from NADPH to NADH. Proc. Natl. Acad. Sci. USA 110, 10946–10951 (2013).
Wong, S. H., Lonhienne, T. G., Winzor, D. J., Schenk, G. & Guddat, L. W. Bacterial and plant ketol-acid reductoisomerases have different mechanisms of induced fit during the catalytic cycle. J. Mol. Biol. 424, 168–179 (2012).
Fraisier, V., Gojon, A., Tillard, P. & Vedele, F. D. Constitutive expression of a putative high-affinity nitrate transporter in Nicotiana plumbaginifolia: evidence for post-transcriptional regulation by a reduced nitrogen source. Plant J. 23, 489–496 (2000).
Hayami, N. & Yamamoto, Y. Y. Primary metabolism and transcriptional regulation in higher plants. Rev. Agric. Sci. 9, 117–127 (2021).
Afitlhile, M., Fukushige, H., Nishimura, M. & Hildebrand, D. F. A defect in glyoxysomal fatty acid β-oxidation reduces jasmonic acid accumulation in Arabidopsis. Plant Physiol. Biochem. 43, 603–609 (2005).
Wasternack, C. & Song, S. Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68, 1303–1321 (2017).
Guo, D. et al. The jasmonate pathway promotes nodule symbiosis and suppresses host plant defense in Medicago truncatula. Mol. Plant 17, 1183–1203 (2024).
Kaur, S., Campbell, B. J. & Suseela, V. Root metabolome of plant–arbuscular mycorrhizal symbiosis mirrors the mutualistic or parasitic mycorrhizal phenotype. N. Phytol. 234, 672–687 (2022).
Akram, N. A., Shafiq, F. & Ashraf, M. Ascorbic acid-A potential oxidant scavenger and its role in plant development and abiotic stress tolerance. Front. Plant Sci. 8, 613 (2017).
Miret, J. A. & Müller, M. AsA/DHA redox pair influencing plant growth and stress tolerance. in (eds Hossain, M. A. et al.) Ascorbic Acid in Plant Growth, Development and Stress Tolerance, 297–319 (Springer International Publishing, 2017).
Belmondo, S. et al. NADPH oxidases in the arbuscular mycorrhizal symbiosis. Plant Signal. Behav. 11, e1165379 (2016).
Fonseca-García, C. et al. Transcriptome analysis of the differential effect of the NADPH oxidase gene RbohB in Phaseolus vulgaris roots following Rhizobium tropici and Rhizophagus irregularis inoculation. BMC Genom. 20, 800 (2019).
Arvola, R., Abshire, E., Bohn, J. & Goldstrohm, A. C. Mechanisms of post-transcriptional gene regulation. in (eds Menon, P. K. M. J. & Goldstrohm, P. A.) Post-transcriptional Mechanisms in Endocrine Regulation, 1–36 (Springer International Publishing, 2016).
Ruiz-Lozano, J. M. et al. Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 39, 441–452 (2016).
Wang, Y. & Wu, Q. S. Influence of sugar metabolism on the dialogue between arbuscular mycorrhizal fungi and plants. Hortic. Adv. 1, 0–0 (2023).
Li, H. et al. Physio-biochemical and transcriptomic features of arbuscular mycorrhizal fungi relieving cadmium stress in wheat. Antioxidants 11, 2390 (2022).
Mishra, A. K. et al. Potentials and prospects of AMF for soil carbon sequestration and nutrient cycling in rice-based cropping system. in (eds Parihar, M., Rakshit, A., Adholeya, A. & Chen, Y.) Arbuscular Mycorrhizal Fungi in Sustainable Agriculture: Nutrient and Crop Management, 113–129 (Springer Nature, 2024).
Wu, J. et al. Arbuscular mycorrhiza augments aluminum tolerance in white clover (Trifolium repens L.) by strengthening the ascorbate-glutathione cycle and phosphorus acquisition. Physiol. Mol. Biol. Plants 29, 1647–1661 (2023).
Scheublin, T. R., Sanders, I. R., Keel, C. & van der Meer, J. R. Characterisation of microbial communities colonising the hyphal surfaces of arbuscular mycorrhizal fungi. ISME J. 4, 752–763 (2010).
Marschner, P. & Baumann, K. Changes in bacterial community structure induced by mycorrhizal colonisation in split-root maize. Plant Soil. 251, 279-289 (2003).
Chen, E., Liao, H., Chen, B. & Shaolin, P. Arbuscular mycorrhizal fungi are a double-edged sword in plant invasion controlled by phosphorus concentration. N. Phytol. 226, 295–300 (2020).
He, D. et al. Flavonoid-attracted Aeromonas sp. from the Arabidopsis root microbiome enhances plant dehydration resistance. ISME J. 16, 2622–2632 (2022).
Abdel-Lateif, K., Bogusz, D. & Hocher, V. The role of flavonoids in the establishment of plant roots endosymbioses with arbuscular mycorrhiza fungi, rhizobia and Frankia bacteria. Plant Signal. Behav. 7, 636–641 (2012).
Besseau, S. et al. Flavonoid accumulation in arabidopsis repressed in lignin synthesis affects auxin transport and plant growth. Plant Cell 19, 148–162 (2007).
Brown, D. E. et al. Flavonoids act as negative regulators of auxin transport in vivo in Arabidopsis. Plant Physiol. 126, 524–535 (2001).
Ji, Z., Belfield, E. J., Li, S., Fu, X. D. & Harberd, N. P. Discovery of a second-site nia2 mutation in the background of multiple ArabidopsisPIF-related mutants containing the pif3-3 allele. N. Phytol. 241, 17–23 (2023).
Medici, A. et al. AtNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 6, 6274 (2015).
Kun, Y. et al. Low phosphorus promotes NSP1–NSP2 heterodimerization to enhance strigolactone biosynthesis and regulate shoot and root architecture in rice. Mol. Plant 16, 1811–1831 (2023).
Singh, S., Katzer, K., Lambert, J., Cerri, M. & Parniske, M. CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152 (2014).
Tatebe, H. & Shiozaki, K. Chapter 91 - Protein serine/threonine-phosphatase 2C (PP2C). in (eds Bradshaw, R. A. & Dennis, E. A.) Handbook of Cell Signaling (Second Edition), 711–716 (Academic Press, 2010).
Wang, J., Munyampundu, J., Xu, Y. & Cai, X. Phylogeny of plant calcium and calmodulin-dependent protein kinases (CCaMKs) and functional analyses of tomato CCaMK in disease resistance. Front. Plant Sci. 6, 1075 (2015).
Sharma, A., Jain, K. K., Jain, A., Kidwai, M. & Kuhad, R. C. Bifunctional in vivo role of laccase exploited in multiple biotechnological applications. Appl. Microbiol Biot. 102, 10327–10343 (2018).
Strong, P. J. & And Claus, H. Laccase: a review of its past and its future in bioremediation. Crit. Rev. Environ. Sci. Tec. 41, 373–434 (2011).
Bell, C. A., Magkourilou, E., Ault, J. R., Urwin, P. E. & Field, K. J. Phytophagy impacts the quality and quantity of plant carbon resources acquired by mutualistic arbuscular mycorrhizal fungi. Nat. Commun. 15, 801 (2024).
Jin, H. et al. The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. N. Phytol. 168, 687–696 (2005).
Dong, S. Study on the effects of different nitrogen, phosphorus, and potassium fertilizer ratios on sugarcane yield and quality. Sugarcane Ind. 000, 16–18 (2007).
Chen, Y. Study on nitrogen fertilizer requirements and optimal application periods for seed sugarcane. Guangxi Sugar Ind. 000, 47–49 (2003).
Wang, Y., Zhang, S. & Zhang, M. Arbuscular mycorrhizal fungal resources and germplasm resources in China (China Agriculture Press, 2012).
Xu, S. GRZJ method to stain arbuscular mycorrhiza fungi in the roots of Guangxi Camellia nitidissima. Chin. J. Trop. Crops 45, 215–224 (2024).
Trouvelot, A., Kough, J. L. & Gianinazzi-Pearson, V. Mesure du taux de mycorhization VA d’un système radiculaire. Recherche de méthodes d’estimation ayant une signification fonctionnelle. in (eds Pearson, G.- V. & Gianinazzi, S.) Physiological and Genetical Aspects of Mycorrhizae (INRA Press, 1986).
Bao, S. Soil agricultural chemical analysis, 3rd edn (China Agricultural Press, 2000).
Kakhki, M. P. & Heidary, M. TRIzol-based RNA extraction: a reliable method for gene expression studies (University of Tehran, 2014).
Souza, G. M. et al. Supporting data for Assembly of the 373K gene space of the polyploid sugarcane genome reveals reservoirsof functional diversity in the world’s leading biomass crop. Gigascience. 8(12), giz129 (2019).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Kim, D. et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14, R36 (2013).
Pertea et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Li, J. et al. TMTpro reagents: a set of isobaric labeling mass tags enables simultaneous proteome-wide measurements across 16 samples. Nat. Methods 17, 399–404 (2020).
Kenneth, J. L., & Thomas, D. S. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT. Methods 25, 402–408 (2001).
Liu, Q. et al. Bio-fertilizer affects structural dynamics, function, and network patterns of the sugarcane rhizospheric microbiota. Micro. Ecol. 84, 1195–1211 (2022).
Wang, J. et al. Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics 12, 116 (2016).
Han, C. et al. Majorbio Cloud 2024: update single-cell and multiomics workflows. iMeta 3, e217 (2024).
Acknowledgements
This work was supported by the Guangxi Science and Technology Major Special Project (Guike AA22117004).
Author information
Authors and Affiliations
Contributions
Q.L., L.M., Z.P., B.C., and Z.Y. designed and managed the project. Q.L., L.M., Z.P. and N.F. collected samples and performed experiments. Q.L., L.M., Y.S., and Z.Y. performed data analyses. Q.L., L.M., and Z.P. wrote the manuscript. N.F., B.C., Y.S., and Z.Y. revised the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Q., Mo, L., Shen, Y. et al. Nitrogen starvation induces arbuscular mycorrhizal fungi to optimize resource allocation in sugarcane roots via suppression of basal metabolism. npj Biofilms Microbiomes (2026). https://doi.org/10.1038/s41522-026-00927-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-026-00927-7