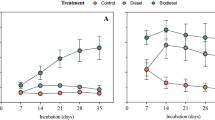
Abstract
The increased use of biodiesel is expected to lead to more microbial corrosion, fouling and fuel degradation issues. In this context, we have analysed the metal, fuel and microbiology of a fouled diesel tank which had been in service for over 30 years. The fuel itself, a B7 biodiesel blend, was not degraded, and—although no free water phase was visible—contained a water content of ~60 ppm. The microbial community was dominated by the fungus Amorphotheca resinae, which formed thick, patchy biofilms on the tank bottom and walls. The tank sheets, composed of galvanised carbon steel, were locally corroded underneath the biofilms, up to a depth of a third of the sheet thickness. On the biofilm-free surfaces, Zn coatings could still be observed. Taken together, A. resinae was shown to thrive in these water-poor conditions, likely enhancing corrosion through the removal of the protective Zn coatings.
Similar content being viewed by others
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information].
References
Söhngen, N. Benzin, petroleum, paraffinöl und paraffin als kohlenstoff-und energiequelle für mikroben. Zentr. Bakteriol. Parasitenk. Abt. II 37, 595–609 (1913).
Bento, F. M. & Gaylarde, C. C. Biodeterioration of stored diesel oil: studies in Brazil. Int. Biodeterior. Biodegrad. 47, 107–112, https://doi.org/10.1016/S0964-8305(00)00112-8 (2001).
Edmonds, P. & Cooney, J. J. Identification of microorganisms isolated from jet fuel systems. Appl. Microbiol. 15, 411–416, https://doi.org/10.1128/am.15.2.411-416.1967 (1967).
Komariah, L. N. et al. Microbial contamination of diesel-biodiesel blends in storage tank; an analysis of colony morphology. Heliyon 8, e09264, https://doi.org/10.1016/j.heliyon.2022.e09264 (2022).
Sørensen, G., Pedersen, D. V., Nørgaard, A. K., Sørensen, K. B. & Nygaard, S. D. Microbial growth studies in biodiesel blends. Bioresour. Technol. 102, 5259–5264, https://doi.org/10.1016/j.biortech.2011.02.017 (2011).
Schleicher, T., Werkmeister, R., Russ, W. & Meyer-Pittroff, R. Microbiological stability of biodiesel–diesel-mixtures. Bioresour. Technol. 100, 724–730, https://doi.org/10.1016/j.biortech.2008.07.029 (2009).
da Fonseca, M. M. B. et al. Unlocking and functional profiling of the bacterial communities in diesel tanks upon additive treatment. Fuel 236, 1311–1320, https://doi.org/10.1016/j.fuel.2018.09.107 (2019).
Marks, C. R. et al. An integrated metagenomic and metabolite profiling study of hydrocarbon biodegradation and corrosion in navy ships. npj Mater. Degrad. 5, 60, https://doi.org/10.1038/s41529-021-00207-z (2021).
Gaylarde, C. C., Bento, F. M. & Kelley, J. Microbial contamination of stored hydrocarbon fuels and its control. Rev. Microbiol. 30, 01–10, https://doi.org/10.1590/S0001-37141999000100001 (1999).
Zobell, C. E. Action of microorganisms on hydrocarbons. Bacteriol. Rev. 10, 1–49 (1946).
Lansdown, A. R. Microbiological attack in aircraft fuel systems. J. R. Aeronautical Soc. 69, 763–767, https://doi.org/10.1017/S0368393100081694 (1965).
Passman, F. J. Microbial contamination and its control in fuels and fuel systems since 1980 – a review. Int. Biodeterior. Biodegrad. 81, 88–104, https://doi.org/10.1016/j.ibiod.2012.08.002 (2013).
Aktas, D. F. et al. Anaerobic hydrocarbon biodegradation and biocorrosion of carbon steel in marine environments: The impact of different ultra low sulfur diesels and bioaugmentation. Int. Biodeterior. Biodegrad. 118, 45–56, https://doi.org/10.1016/j.ibiod.2016.12.013 (2017).
Bento, F. M., Beech, I. B., Gaylarde, C. C., Englert, G. E. & Muller, I. L. Degradation and corrosive activities of fungi in a diesel–mild steel–aqueous system. World J. Microbiol. Biotechnol. 21, 135–142, https://doi.org/10.1007/s11274-004-3042-2 (2005).
Floyd, J. G., Stamps, B. W., Goodson, W. J. & Stevenson, B. S. Locating and quantifying carbon steel corrosion rates linked to fungal B20 biodiesel degradation. Appl. Environ. Microbiol. 87, e0117721, https://doi.org/10.1128/aem.01177-21 (2021).
Ching, T. H. et al. Biodegradation of biodiesel and microbiologically induced corrosion of 1018 steel by Moniliella wahieum Y12. Int. Biodeterior. Biodegrad. 108, 122–126, https://doi.org/10.1016/j.ibiod.2015.11.027 (2016).
Aslan, C., Aulia, N. I., Devianto, H. & Harimawan, A. Influence of axenic culture of Bacillus clausii and mixed culture on biofilm formation, carbon steel corrosion, and methyl ester degradation in B30 storage tank system. J. Environ. Chem. Eng. 10, 108013, https://doi.org/10.1016/j.jece.2022.108013 (2022).
Xu, D., Gu, T. & Lovley, D. R. Microbially mediated metal corrosion. Nat. Rev. Microbiol. 21, 705–718, https://doi.org/10.1038/s41579-023-00920-3 (2023).
Knisz, J. et al. Microbiologically influenced corrosion—more than just microorganisms. FEMS Microbiol. Rev. 47, fuad041 (2023).
Khan, S. et al. Harnessing bacterial power: advanced strategies and genetic engineering insights for biocorrosion control and inhibition. npj Mater. Degrad. 9, 74, https://doi.org/10.1038/s41529-025-00623-5 (2025).
Luz, G. V. S., Sousa, B. A. S. M., Guedes, A. V., Barreto, C. C. & Brasil, L. M. Biocides used as additives to biodiesels and their risks to the environment and public health: a review. Molecules 23, 2698, https://doi.org/10.3390/molecules23102698 (2018).
Bailey, C. A. & May, M. E. Evaluation of microbiological test kits for hydrocarbon fuel systems. Appl. Environ. Microbiol. 37, 871–877, https://doi.org/10.1128/aem.37.5.871-877.1979 (1979).
Lopes, P. T. C. & Gaylarde, C. Use of immunofluorescence to detect the fungus Hormoconis resinae in aviation kerosine. Int. Biodeterior. Biodegrad. 37, 37–40, https://doi.org/10.1016/0964-8305(95)00110-7 (1996).
Martin-Sanchez, P. M., Gorbushina, A. A., Kunte, H. J. & Toepel, J. A novel qPCR protocol for the specific detection and quantification of the fuel-deteriorating fungus Hormoconis resinae. Biofouling 32, 635–644, https://doi.org/10.1080/08927014.2016.1177515 (2016).
Bücker, F. et al. Evaluation of the deteriogenic microbial community using qPCR, n-alkanes and FAMEs biodegradation in diesel, biodiesel and blends (B5, B10, and B50) during storage. Fuel 233, 911–917, https://doi.org/10.1016/j.fuel.2017.11.076 (2018).
Amsellem, D. et al. Perspectives d’évolution des biocarburants: jeux des acteurs et enjeux fonciers. (L’Observatoire de la sécurité des flux et des matières énergétiques, 2021).
He, B. B., Thompson, J. C., Routt, D. W. & Van Gerpen, J. H. Moisture absorption in biodiesel and its petro-diesel blends. Appl. Eng. Agric. 23, 6, https://doi.org/10.13031/2013.22320 (2007).
Dodos, G., Konstantakos, T., Longinos, S. & Zannikos, F. Effects of microbiological contamination in the quality of biodiesel fuels. Glob. NEST J. 14, 175–182, https://doi.org/10.30955/gnj.000856 (2012).
Lee, J. S., Ray, R. I. & Little, B. J. An assessment of alternative diesel fuels: microbiological contamination and corrosion under storage conditions. Biofouling 26, 623–635, https://doi.org/10.1080/08927014.2010.504984 (2010).
Lyles, C. N. et al. Impact of organosulfur content on diesel fuel stability and implications for carbon steel corrosion. Environ. Sci. Technol. 47, 6052–6062, https://doi.org/10.1021/es4006702 (2013).
EPA, U. Investigation Of Corrosion-Influencing Factors In Underground Storage Tanks With Diesel Service. EPA 510-R-16-001. (United States Environmental Protection Agency, 2016).
Houbraken, J., Visagie Cobus, M. & Frisvad Jens, C. Recommendations to prevent taxonomic misidentification of genome-sequenced fungal strains. Microbiol. Resour. Announc. 10, e01074–01020, https://doi.org/10.1128/MRA.01074-20 (2021).
Stamps, B. W. et al. In situ linkage of fungal and bacterial proliferation to microbiologically influenced corrosion in B20 biodiesel storage tanks. Front. Microbiol. 11, 167, https://doi.org/10.3389/fmicb.2020.00167 (2020).
Cofone, L., Walker, J. D. & Cooney, J. J. Utilization of hydrocarbons by Cladosporium resinae. Microbiology 76, 243–246, https://doi.org/10.1099/00221287-76-1-243 (1973).
Lindley, N. D. & Heydeman, M. T. The uptake of n-alkanes from alkane mixtures during growth of the hydrocarbon-utilizing fungus Cladosporium resinae. Appl. Microbiol. Biotechnol. 23, 384–388, https://doi.org/10.1007/BF00257038 (1986).
Krivushina, A. A., Bobyreva, T. V. & Mokeeva, V. L. Growth ability of “kerosene fungus” Amorphotheca resinae strains isolated from different habitats in aviation fuel. Biol. Bull. Rev. 13, S93–S98, https://doi.org/10.1134/S2079086423070095 (2023).
Parbery, D. G. The Kerosene Fungus, Amorphotheca resinae: Its Biology, Taxonomy and Control. (University of Melbourne, 1970).
Krohn, I. et al. Deep (meta)genomics and (meta)transcriptome analyses of fungal and bacteria consortia from aircraft tanks and kerosene identify key genes in fuel and tank corrosion. Front. Microbiol. 12, 722259 (2021).
Walker, J. D. & Cooney, J. J. Aliphatic hydrocarbons of Cladosporium resinae cultured on glucose, glutamic acid, and hydrocarbons. Appl. Microbiol. 26, 705–708, https://doi.org/10.1128/am.26.5.705-708.1973 (1973).
Schirmer, M. et al. Insight into biases and sequencing errors for amplicon sequencing with the Illumina MiSeq platform. Nucleic Acids Res. 43, e37–e37, https://doi.org/10.1093/nar/gku1341 (2015).
Martino, E. et al. Comparative genomics and transcriptomics depict ericoid mycorrhizal fungi as versatile saprotrophs and plant mutualists. N. Phytol. 217, 1213–1229, https://doi.org/10.1111/nph.14974 (2018).
Wang, W., Jenkins, P. E. & Ren, Z. Heterogeneous corrosion behaviour of carbon steel in water contaminated biodiesel. Corros. Sci. 53, 845–849, https://doi.org/10.1016/j.corsci.2010.10.020 (2011).
Fang, H. L. & McCormick, R. L. Spectroscopic study of biodiesel degradation pathways. SAE Technical Paper 2006-01-3300 (2006).
Bücker, F. et al. Impact of biodiesel on biodeterioration of stored Brazilian diesel oil. Int. Biodeterior. Biodegrad. 65, 172–178, https://doi.org/10.1016/j.ibiod.2010.09.008 (2011).
Marder, A. R. The metallurgy of zinc-coated steel. Prog. Mater. Sci. 45, 191–271, https://doi.org/10.1016/S0079-6425(98)00006-1 (2000).
Ku, Y.-Y., Tang, T.-W., Lin, K. W. & Chan, S. The impact upon applicability of metal fuel tank using different biodiesel. SAE Int. J. Mater. Manuf. 8, 757–764, https://doi.org/10.4271/2015-01-0521 (2015).
Ackermann, H. et al. Entwicklung einer Prüfmethode zur Bewertung der Materialbeständigkeit von Bauteilen in Mitteldestillatanwendungen. (DGMK, 2020).
Fernandes, D. M. et al. Storage stability and corrosive character of stabilised biodiesel exposed to carbon and galvanised steels. Fuel 107, 609–614, https://doi.org/10.1016/j.fuel.2012.11.010 (2013).
Mackowiak, J. & Short, N. R. Metallurgy of galvanized coatings. Int. Met. Rev. 24, 1–19, https://doi.org/10.1179/imtr.1979.24.1.1 (1979).
Townsend, H. & Hart, R. Composition of chromate passivation films on aluminum-zinc alloy-coated sheet steel. J. Electrochem. Soc. 131, 1345–1348, https://doi.org/10.1149/1.2115817 (1984).
Buzzini, P., Turchetti, B. & Yurkov, A. Extremophilic yeasts: the toughest yeasts around? Yeast 35, 487–497, https://doi.org/10.1002/yea.3314 (2018).
Deveau, A. et al. Bacterial–fungal interactions: ecology, mechanisms and challenges. FEMS Microbiol. Rev. 42, 335–352, https://doi.org/10.1093/femsre/fuy008 (2018).
Alexander, A., Singh, V. K. & Mishra, A. Halotolerant PGPR Stenotrophomonas maltophilia BJ01 induces salt tolerance by modulating physiology and biochemical activities of Arachis hypogaea. Front. Microbiol. 11, 568289 (2020).
Belyakova, E. V. et al. The new facultatively chemolithoautotrophic, moderately halophilic, sulfate-reducing bacterium Desulfovermiculus halophilus gen. nov., sp. nov., isolated from an oil field. Microbiology 75, 161–171, https://doi.org/10.1134/S0026261706020093 (2006).
Oren, A. Thermodynamic limits to microbial life at high salt concentrations. Environ. Microbiol. 13, 1908–1923, https://doi.org/10.1111/j.1462-2920.2010.02365.x (2011).
Steinle, L. et al. Life on the edge: active microbial communities in the Kryos MgCl(2)-brine basin at very low water activity. Isme J. 12, 1414–1426, https://doi.org/10.1038/s41396-018-0107-z (2018).
Ryan, R. P. et al. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat. Rev. Microbiol. 7, 514–525, https://doi.org/10.1038/nrmicro2163 (2009).
Shapiro, T. et al. Revealing of non-cultivable bacteria associated with the mycelium of fungi in the kerosene-degrading community isolated from the contaminated jet fuel. J. Fungi 7, 43, https://doi.org/10.3390/jof7010043 (2021).
Gunasekera, T. S., Striebich, R. C., Mueller, S. S., Strobel, E. M. & Ruiz, O. N. Transcriptional profiling suggests that multiple metabolic adaptations are required for effective proliferation of Pseudomonas aeruginosa in jet fuel. Environ. Sci. Technol. 47, 13449–13458, https://doi.org/10.1021/es403163k (2013).
Martin-Sanchez, P. M., Gorbushina, A. A. & Toepel, J. Quantification of microbial load in diesel storage tanks using culture- and qPCR-based approaches. Int. Biodeterior. Biodegrad. 126, 216–223, https://doi.org/10.1016/j.ibiod.2016.04.009 (2018).
Prenafeta-Boldú, F. X., de Hoog, G. S. & Summerbell, R. C. in Microbial Communities Utilizing Hydrocarbons and Lipids (ed McGenity, T. J.) 307–342 (Springer, 2019).
Hill, E. & Thomas, A. in Proceedings of the Third International Biodegradation Symposium 157–174 (1977).
van Beilen, J. B. & Funhoff, E. G. Alkane hydroxylases involved in microbial alkane degradation. Appl. Microbiol. Biotechnol. 74, 13–21, https://doi.org/10.1007/s00253-006-0748-0 (2007).
Gunde-Cimerman, N., Plemenitaš, A. & Oren, A. Strategies of adaptation of microorganisms of the three domains of life to high salt concentrations. FEMS Microbiol. Rev. 42, 353–375, https://doi.org/10.1093/femsre/fuy009 (2018).
Hettige, G. E. G. & Sheridan, J. E. Interactions of fungi contaminating diesel fuel. Int. Biodeterior. 25, 299–309, https://doi.org/10.1016/0265-3036(89)90004-3 (1989).
Gerwien, F., Skrahina, V., Kasper, L., Hube, B. & Brunke, S. Metals in fungal virulence. FEMS Microbiol. Rev. 42, fux050 (2017).
Lonergan, Z. R. & Skaar, E. P. Nutrient zinc at the host–pathogen interface. Trends Biochem. Sci. 44, 1041–1056, https://doi.org/10.1016/j.tibs.2019.06.010 (2019).
Robinson, J. R., Isikhuemhen, O. S. & Anike, F. N. Fungal–metal interactions: a review of toxicity and homeostasis. J. Fungi 7, 225, https://doi.org/10.3390/jof7030225 (2021).
McDevitt, C. A. et al. A molecular mechanism for bacterial susceptibility to zinc. PLOS Pathog. 7, e1002357, https://doi.org/10.1371/journal.ppat.1002357 (2011).
Simm, C. et al. Saccharomyces cerevisiae vacuole in zinc storage and intracellular zinc distribution. Eukaryot. Cell 6, 1166–1177, https://doi.org/10.1128/ec.00077-07 (2007).
Cho, M. et al. Vacuolar zinc transporter Zrc1 is required for detoxification of excess intracellular zinc in the human fungal pathogen Cryptococcus neoformans. J. Microbiol. 56, 65–71, https://doi.org/10.1007/s12275-018-7475-y (2018).
Ruytinx, J. et al. Zinc export results in adaptive zinc tolerance in the ectomycorrhizal basidiomycete Suillus bovinus. Metallomics 5, 1225–1233, https://doi.org/10.1039/c3mt00061c (2013).
Leonhardt, T., Sácký, J., Šimek, P., Šantrůček, J. & Kotrba, P. Metallothionein-like peptides involved in sequestration of Zn in the Zn-accumulating ectomycorrhizal fungus Russula atropurpurea. Metallomics 6, 1693–1701, https://doi.org/10.1039/c4mt00141a (2014).
Kalsotra, T., Khullar, S., Agnihotri, R. & Reddy, M. S. Metal induction of two metallothionein genes in the ectomycorrhizal fungus Suillus himalayensis and their role in metal tolerance. Microbiology 164, 868–876, https://doi.org/10.1099/mic.0.000666 (2018).
Fomina, M., Charnock, J., Bowen, A. D. & Gadd, G. M. X-ray absorption spectroscopy (XAS) of toxic metal mineral transformations by fungi. Environ. Microbiol. 9, 308–321, https://doi.org/10.1111/j.1462-2920.2006.01139.x (2007).
Kleijburg, F. E. L., Safeer, A. A., Baldus, M. & Wösten, H. A. B. Binding of micro-nutrients to the cell wall of the fungus Schizophyllum commune. Cell Surf. 10, 100108, https://doi.org/10.1016/j.tcsw.2023.100108 (2023).
San-Blas, G., Guanipa, O., Moreno, B., Pekerar, S. & San-Blas, F. Cladosporium carrionii and Hormoconis resinae (C. resinae): Cell wall and melanin studies. Curr. Microbiol. 32, 11–16, https://doi.org/10.1007/s002849900003 (1996).
Fogarty, R. V. & Tobin, J. M. Fungal melanins and their interactions with metals. Enzym. Microb. Technol. 19, 311–317, https://doi.org/10.1016/0141-0229(96)00002-6 (1996).
Gadd, G. M. & Derome, L. Biosorption of copper by fungal melanin. Appl. Microbiol. Biotechnol. 29, 610–617, https://doi.org/10.1007/BF00260993 (1988).
Juzeliūnas, E. et al. Microbially influenced corrosion of zinc and aluminium – two-year subjection to influence of Aspergillus niger. Corros. Sci. 49, 4098–4112, https://doi.org/10.1016/j.corsci.2007.05.004 (2007).
Koblitz, J. et al. MediaDive: the expert-curated cultivation media database. Nucleic Acids Res. 51, D1531–D1538, https://doi.org/10.1093/nar/gkac803 (2022).
Rachel, N. M. & Gieg, L. M. Preserving microbial community integrity in oilfield produced water. Front. Microbiol. 11, 581387 (2020).
Tedersoo, L. et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 10, 4852 (2015).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583, https://doi.org/10.1038/nmeth.3869 (2016).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17, 3, https://doi.org/10.14806/ej.17.1.200 (2011).
Quast, C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596, https://doi.org/10.1093/nar/gks1219 (2012).
Abarenkov, K. et al. The UNITE database for molecular identification and taxonomic communication of fungi and other eukaryotes: sequences, taxa and classifications reconsidered. Nucleic Acids Res. 52, 791–797, https://doi.org/10.1093/nar/gkad1039 (2023).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. Plos One 8, e61217, https://doi.org/10.1371/journal.pone.0061217 (2013).
Alibrandi, A., di Primio, R., Bartholomäus, A. & Kallmeyer, J. A modified isooctane-based DNA extraction method from crude oil. mLife 2, 328–338, https://doi.org/10.1002/mlf2.12081 (2023).
Kieser, S., Brown, J., Zdobnov, E. M., Trajkovski, M. & McCue, L. A. ATLAS: a snakemake workflow for assembly, annotation, and genomic binning of metagenome sequence data. BMC Bioinform. 21, 257, https://doi.org/10.1186/s12859-020-03585-4 (2020).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359, https://doi.org/10.1038/nmeth.1923 (2012).
Curry, K. D. et al. Emu: species-level microbial community profiling of full-length 16S rRNA Oxford Nanopore sequencing data. Nat. Methods 19, 845–853, https://doi.org/10.1038/s41592-022-01520-4 (2022).
Camacho, C. et al. BLAST+: architecture and applications. BMC Bioinform. 10, 421, https://doi.org/10.1186/1471-2105-10-421 (2009).
Spurr, A. R. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26, 31–43, https://doi.org/10.1016/S0022-5320(69)90033-1 (1969).
DIN. EN ISO/IEC 17025:2018-03, General requirements for the competence of testing and calibration laboratories. (2018).
Acknowledgements
This study was supported by internal funds of the B.A.M., with amplicon sequencing supported by NSERC Discovery and Genome Canada grants awarded to L.M.G. We thank Alexander Bartholomäus of GreenGate Genomics for the metagenomic whole genome analysis.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
R.G.: sampling and sample preparation, data acquisition, curation, visualisation, writing original draft; A.Y.: analysis of fungal SEM images and sequencing data; I.F. conducted ESEM and EDX analyses; M.D., U.K.: technical support, sample preparation; M.N.S., J.W., S.K.,B.A.S.: aseptic sampling, isolation of prokaryotes; Ra.Bä.: corrosion methodology and data analysis; R.H., A.H., M.W., T.M., H.S., Y.S., N.M.T., K.L.G.: sample treatment and data acquisition; Ro.Be.: diesel expertise, data analysis; O.O.: conceptualisation, corrosion analysis; J.G.: detection of the corrosion case, provision of the fuel tank for study; supplying comprehensive information on the tank failure and thevehicle's 30-year operational history; J.S.: analysis of fungal biology and genomes; A.A.G.: conceptualisation; sampling, SEM analysis,funding acquisition; L.M.G.: conceptualisation, sampling, SEM analysis, writing original draft, funding acquisition. Writing – review & editing: all co-authors.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Gerrits, R., Stepec, B.A., Bäßler, R. et al. A 30-year-old diesel tank: fungal-dominated biofilms cause local corrosion of galvanised steel. npj Mater Degrad (2026). https://doi.org/10.1038/s41529-025-00731-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41529-025-00731-2