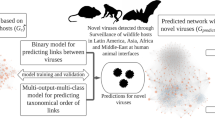
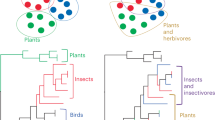
Abstract
Better methods to predict and prevent the emergence of zoonotic viruses could support future efforts to reduce the risk of epidemics. We propose a network science framework for understanding and predicting human and animal susceptibility to viral infections. Related approaches have so far helped to identify basic biological rules that govern cross-species transmission and structure the global virome. We highlight ways to make modelling both accurate and actionable, and discuss the barriers that prevent researchers from translating viral ecology into public health policies that could prevent future pandemics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Jones, K. E. et al. Global trends in emerging infectious diseases. Nature 451, 990–993 (2008).
Woolhouse, M. E. et al. Temporal trends in the discovery of human viruses. Proc. R. Soc. B 275, 2111–2115 (2008).
Smith, K. F. et al. Global rise in human infectious disease outbreaks. J. R. Soc. Interface 11, 20140950 (2014).
Carlson, C. J. et al. Climate change will drive novel cross-species viral transmission. Preprint at bioRxiv https://doi.org/10.1101/2020.01.24.918755 (2020).
Swei, A., Couper, L. I., Coffey, L. L., Kapan, D. & Bennett, S. Patterns, drivers, and challenges of vector-borne disease emergence. Vector Borne Zoonotic Dis. 20, 159–170 (2020).
Belay, E. D. et al. Zoonotic disease programs for enhancing global health security. Emerg. Infect. Dis. 23, S65 (2017).
Morse, S. S. et al. Prediction and prevention of the next pandemic zoonosis. Lancet 380, 1956–1965 (2012).
Carroll, D. et al. The global virome project. Science 359, 872–874 (2018).
Carlson, C. J., Zipfel, C. M., Garnier, R. & Bansal, S. Global estimates of mammalian viral diversity accounting for host sharing. Nat. Ecol. Evol. 3, 1070–1075 (2019).
Babayan, S. A., Orton, R. J. & Streicker, D. G. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science 362, 577–580 (2018).
Han, B. A. et al. Undiscovered bat hosts of filoviruses. PLoS Negl. Trop. Dis. 10, e0004815 (2016).
Schmidt, J. P. et al. Spatiotemporal fluctuations and triggers of Ebola virus spillover. Emerg. Infect. Dis. 23, 415 (2017).
Guth, S., Visher, E., Boots, M. & Brook, C. E. Host phylogenetic distance drives trends in virus virulence and transmissibility across the animal–human interface. Phil. Trans. R. Soc. Biol. Sci. 374, 20190296 (2019).
Glennon, E. E. et al. Syndromic detectability of haemorrhagic fever outbreaks. Preprint at medRxiv https://doi.org/10.1101/2020.03.28.20019463 (2020).
Pigott, D. M. et al. Local, national, and regional viral haemorrhagic fever pandemic potential in Africa: a multistage analysis. Lancet 390, 2662–2672 (2017).
Palmer, S., Brown, D. & Morgan, D. Early qualitative risk assessment of the emerging zoonotic potential of animal diseases. BMJ 331, 1256–1260 (2005).
Grange, Z. L. et al. Ranking the risk of animal-to-human spillover for newly discovered viruses. Proc. Natl Acad. Sci. USA 118, e2002324118 (2021).
Carlson, C. J. From PREDICT to prevention, one pandemic later. Lancet Microbe 1, e6–e7 (2020).
Holmes, E., Rambaut, A. & Andersen, K. Pandemics: spend on surveillance, not prediction. Nature 558, 180–182 (2018).
Breiman, L. Statistical modeling: the two cultures (with comments and a rejoinder by the author). Stat. Sci. 16, 199–231 (2001).
Mouquet, N. et al. Predictive ecology in a changing world. J. Appl. Ecol. 52, 1293–1310 (2015).
Olival, K. J. et al. Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017).
Stephens, P. R. et al. Global mammal parasite database version 2.0. Ecology 98, 1476 (2017).
Wardeh, M., Risley, C., McIntyre, M. K., Setzkorn, C. & Baylis, M. Database of host–pathogen and related species interactions, and their global distribution. Sci. Data 2, 150049 (2015).
Shaw, L. P. et al. The phylogenetic range of bacterial and viral pathogens of vertebrates. Mol. Ecol. 29, 3361–3379 (2020).
Gibb, R. et al. Data proliferation, reconciliation, and synthesis in viral ecology. BioScience https://doi.org/10.1093/biosci/biab080 (2021).
Dallas, T., Park, A. W. & Drake, J. M. Predicting cryptic links in host–parasite networks. PLoS Comput. Biol. 13, e1005557 (2017).
Poisot, T. et al. Imputing the mammalian virome with linear filtering and singular value decomposition. Preprint at https://arxiv.org/abs/2105.14973 (2021).
Carlson, C. J. et al. The Global Virome in One Network (VIRION): an atlas of vertebrate–virus associations. Preprint at bioRxiv https://doi.org/10.1101/2021.08.06.455442 (2021).
Albery, G. F., Eskew, E. A., Ross, N. & Olival, K. J. Predicting the global mammalian viral sharing network using phylogeography. Nat. Commun. 11, 2260 (2020).
Davies, T. J. & Pedersen, A. B. Phylogeny and geography predict pathogen community similarity in wild primates and humans. Proc. R. Soc. B Biol. Sci. 275, 1695–1701 (2008).
Guy, C., Thiagavel, J., Mideo, N. & Ratcliffe, J. M. Phylogeny matters: revisiting ‘a comparison of bats and rodents as reservoirs of zoonotic viruses’. R. Soc. Open Sci. 6, 181182 (2019).
Washburne, A. D. et al. Taxonomic patterns in the zoonotic potential of mammalian viruses. PeerJ 6, e5979 (2018).
Plowright, R. K. et al. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 15, 502 (2017).
Stephens, P. R. et al. The macroecology of infectious diseases: a new perspective on global-scale drivers of pathogen distributions and impacts. Ecol. Lett. 19, 1159–1171 (2016).
Longdon, B., Brockhurst, M. A., Russell, C. A., Welch, J. J. & Jiggins, F. M. The evolution and genetics of virus host shifts. PLoS Pathog. 10, e1004395 (2014).
Farrell, M. J., Elmasri, M., Stephens, D. A. & Davies, T. J. Predicting missing links in global host–parasite networks. bioRxiv https://doi.org/10.1101/2020.02.25.965046 (2020).
Gilbert, A. T. et al. Deciphering serology to understand the ecology of infectious diseases in wildlife. EcoHealth 10, 298–313 (2013).
Becker, D. J., Seifert, S. N. & Carlson, C. J. Beyond infection: integrating competence into reservoir host prediction. Trends Ecol. Evol. 35, 1062–1065 (2020).
Walsh, M. G., Mor, S. M., Maity, H. & Hossain, S. A preliminary ecological profile of Kyasanur Forest disease virus hosts among the mammalian wildlife of the Western Ghats, India. Ticks Tick Borne Dis. 11, 101419 (2020).
Plowright, R. K. et al. Prioritizing surveillance of Nipah virus in India. PLoS Negl. Trop. Dis. 13, e0007393 (2019).
Schmidt, J. P. et al. Ecological indicators of mammal exposure to Ebolavirus. Philos. Trans. R. Soc. B Biol. Sci. 374, 20180337 (2019).
Worsley-Tonks, K. E. et al. Using host traits to predict reservoir host species of rabies virus. PLoS Negl. Trop. Dis. 14, e0008940 (2020).
Woolhouse, M. E. & Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 11, 1842 (2005).
Johnson, C. K. et al. Spillover and pandemic properties of zoonotic viruses with high host plasticity. Sci. Rep. 5, 14830 (2015).
Elena, S. F. & Sanjuán, R. Adaptive value of high mutation rates of RNA viruses: separating causes from consequences. J. Virol. 79, 11555–11558 (2005).
Duffy, S. Why are RNA virus mutation rates so damn high? PLoS Biol. 16, e3000003 (2018).
Grewelle, R. E. Larger viral genome size facilitates emergence of zoonotic diseases. Preprint at bioRxiv https://doi.org/10.1101/2020.03.10.986109 (2020).
Mollentze, N. & Streicker, D. G. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proc. Natl Acad. Sci. USA 117, 9423–9430 (2020).
Walker, J. W., Han, B. A., Ott, I. M. & Drake, J. M. Transmissibility of emerging viral zoonoses. PLoS ONE 13, e0206926 (2018).
Damas, J. et al. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc. Natl Acad. Sci. USA https://doi.org/10.1073/pnas.2010146117 (2020).
Zhang, Z. et al. Rapid identification of human-infecting viruses. Transbound. Emerg. Dis. 66, 2517–2522 (2019).
Eng, C. L., Tong, J. C. & Tan, T. W. Predicting zoonotic risk of influenza A viruses from host tropism protein signature using random forest. Int. J. Mol. Sci. 18, 1135 (2017).
Li, J. et al. Machine learning methods for predicting human-adaptive influenza A viruses based on viral nucleotide compositions. Mol. Biol. Evol. 37, 1224–1236 (2020).
Kim, B., Niu, X., Hunter, D. R. & Cao, X. A dynamic additive and multiplicative effects model with application to the United Nations voting behaviors. Preprint at https://arxiv.org/abs/1803.06711 (2018).
Becker, D. et al. Optimizing predictive models to prioritize viral discovery in zoonotic reservoirs. Lancet Microbe (in the press).
Han, B. A., Schmidt, J. P., Bowden, S. E. & Drake, J. M. Rodent reservoirs of future zoonotic diseases. Proc. Natl Acad. Sci. USA 112, 7039–7044 (2015).
Plourde, B. T. et al. Are disease reservoirs special? Taxonomic and life history characteristics. PLoS ONE 12, e0180716 (2017).
Keesing, F. et al. Impacts of biodiversity on the emergence and transmission of infectious diseases. Nature 468, 647–652 (2010).
Albery, G. F. & Becker, D. J. Fast-lived hosts and zoonotic risk. Trends Parasitol. 37, 117–129 (2021).
Young, C. C. & Olival, K. J. Optimizing viral discovery in bats. PLoS ONE 11, e0149237 (2016).
Albery, G. F. et al. Urban-adapted mammal species have more known pathogens. Preprint at bioRxiv https://doi.org/10.1101/2021.01.02.425084 (2021).
Wille, M., Geoghegan, J. L. & Holmes, E. C. How accurately can we assess zoonotic risk? PLoS Biol. 19, e3001135 (2021).
Gibb, R. et al. Mammal virus diversity estimates are unstable due to accelerating discovery effort. Preprint at bioRxiv https://doi.org/10.1101/2021.08.10.455791 (2021).
Xu, G. J. et al. Comprehensive serological profiling of human populations using a synthetic human virome. Science 348, aaa0698 (2015).
Geoghegan, J. L. & Holmes, E. C. Predicting virus emergence amid evolutionary noise. Open Biol. 7, 170189 (2017).
Fischhoff, I. R., Castellanos, A. A., Rodrigues, J. P., Varsani, A. & Han, B. A. Predicting the zoonotic capacity of mammals to transmit SARS-CoV-2. Proc. R. Soc. B Biol. Sci. https://doi.org/10.1098/rspb.2021.1651 (2021).
Hou, Y. et al. Angiotensin-converting enzyme 2 (ACE2) proteins of different bat species confer variable susceptibility to SARS-CoV entry. Arch. Virol. 155, 1563–1569 (2010).
Thompson, A. J., de Vries, R. P. & Paulson, J. C. Virus recognition of glycan receptors. Curr. Opin. Virol. 34, 117–129 (2019).
Kocher, J. F. et al. Bat caliciviruses and human noroviruses are antigenically similar and have overlapping histo-blood group antigen binding profiles. Mbio 9, e00869-18 (2018).
Chiramel, A. I. et al. TRIM5α restricts flavivirus replication by targeting the viral protease for proteasomal degradation. Cell Rep. 27, 3269–3283 (2019).
Young, F., Rogers, S. & Robertson, D. L. Predicting host taxonomic information from viral genomes: a comparison of feature representations. PLoS Comput. Biol. 16, e1007894 (2020).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Truong, P., Garcia-Vallve, S. & Puigbo, P. An unsupervised algorithm for host identification in flaviviruses. Life https://doi.org/10.3390/life11050442 (2021).
Mollentze, N., Babayan, S. & Streicker, D. Identifying and prioritizing potential human-infecting viruses from their genome sequences. PLoS Biol. 19, e3001390 (2021).
Wang, W. et al. A network-based integrated framework for predicting virus–prokaryote interactions. NAR Genom. Bioinform. 2, lqaa044 (2020).
Bartoszewicz, J. M., Seidel, A. & Renard, B. Y. Interpretable detection of novel human viruses from genome sequencing data. NAR Genom. Bioinform. 3, lqab004 (2021).
He, X. et al. Neural collaborative filtering. In Proc. 26th International Conference on World Wide Web 26, 173–182 (Republic and Canton of Geneva, Switzerland, 2017).
Fout, A., Byrd, J., Shariat, B. & Ben-Hur, A. Protein interface prediction using graph convolutional networks. NIPS’17: Proc. 31st International Conference on Neural Information Processing Systems 31, 6533–6542 (2017).
Hamilton, W. L., Ying, R. & Leskovec, J. Representation learning on graphs: methods and applications. IEEE Data Eng. Bull. 40, 52–74 (2017).
Bergner, L. M. et al. Characterizing and evaluating the zoonotic potential of novel viruses discovered in vampire bats. Viruses 13, 252 (2021).
Dietze, M. C. et al. Iterative near-term ecological forecasting: needs, opportunities, and challenges. Proc. Natl Acad. Sci. USA 115, 1424–1432 (2018).
Schulz, J. E. et al. Serological evidence for henipa-like and filo-like viruses in Trinidad bats. J. Infect. Dis. 221, S375–S382 (2020).
Brook, C. E. et al. Disentangling serology to elucidate henipa- and filovirus transmission in Madagascar fruit bats. J. Anim. Ecol. 88, 1001–1016 (2019).
Seifert, S. N. et al. Rousettus aegyptiacus bats do not support productive Nipah virus replication. J. Infect. Dis. 221, S407–S413 (2020).
Carlson, C. J. et al. The future of zoonotic risk prediction. Phil. Trans. R. Soc. B Biol. Sci. 376, 20200358 (2021).
Ge, X.-Y. et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature 503, 535–538 (2013).
Menachery, V. D. et al. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat. Med. 21, 1508–1513 (2015).
Guan, Y. et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302, 276–278 (2003).
Woo, P. C. Y. et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79, 884–895 (2005).
Li, W. et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 310, 676–679 (2005).
Wang, M. et al. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 11, 1860–1865 (2005).
Hu, B. et al. Discovery of a rich gene pool of bat SARS-related coronaviruses provides new insights into the origin of SARS coronavirus. PLoS Pathog. 13, e1006698 (2017).
Zhou, P. et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020).
Xiao, K. et al. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583, 286–289 (2020).
Lam, T.-Y. et al. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020).
Wacharapluesadee, S. et al. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat. Commun. 12, 972 (2021).
Holmes, E. C. et al. The origins of SARS-CoV-2: a critical review. Cell 184, 4848–4856 (2021).
Oude Munnink, B. B. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177 (2021).
Chandler, J. C. et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Proc. Natl Acad. Sci. USA 118, e2114828118 (2021).
Jia, P., Dai, S., Wu, T. & Yang, S. New approaches to anticipate the risk of reverse zoonosis. Trends Ecol. Evol. 36, 580–590 (2021).
Lednicky, J. A. et al. Isolation of a novel recombinant canine coronavirus from a visitor to Haiti: further evidence of transmission of coronaviruses of zoonotic origin to humans. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciab924 (2021).
Vlasova, A. N. et al. Novel canine coronavirus isolated from a hospitalized pneumonia patient, East Malaysia. Clin. Infect. Dis. https://doi.org/10.1093/cid/ciab456 (2021).
Lednicky, J. A. et al. Emergence of porcine delta-coronavirus pathogenic infections among children in Haiti through independent zoonoses and convergent evolution. Preprint at medRxiv https://doi.org/10.1101/2021.03.19.21253391 (2021).
Hay, A. J. & McCauley, J. W. The WHO global influenza surveillance and response system (GISRS)—a future perspective. Influenza Other Respir. Viruses 12, 551–557 (2018).
Subbarao, K. et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 279, 393–396 (1998).
Kandeel, A. et al. Zoonotic transmission of avian influenza virus (H5N1), Egypt, 2006–2009. Emerg. Infect. Dis. 16, 1101 (2010).
Ke, C. et al. Human infection with highly pathogenic avian influenza A (H7N9) virus, China. Emerg. Infect. Dis. 23, 1332 (2017).
Gaidet, N. et al. Evidence of infection by H5N2 highly pathogenic avian influenza viruses in healthy wild waterfowl. PLoS Pathog. 4, e1000127 (2008).
Webster, R. G., Bean, W. J., Gorman, O. T., Chambers, T. M. & Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Mol. Biol. Rev. 56, 152–179 (1992).
Pawar, S. D. et al. Avian influenza surveillance reveals presence of low pathogenic avian influenza viruses in poultry during 2009–2011 in the West Bengal State, India. Virol. J. 9, 151 (2012).
Parry, R., Wille, M., Turnbull, O. M., Geoghegan, J. L. & Holmes, E. C. Divergent influenza-like viruses of amphibians and fish support an ancient evolutionary association. Viruses 12, 1042 (2020).
Campbell, P. J. et al. The M segment of the 2009 pandemic influenza virus confers increased neuraminidase activity, filamentous morphology, and efficient contact transmissibility to A/Puerto Rico/8/1934-based reassortant viruses. J. Virol. 88, 3802–3814 (2014).
Carlson, C. Evolutionary surprise, artificial intelligence, and H5N8. The Verena Blog https://www.viralemergence.org/blog/evolutionary-surprise-artificial-intelligence-and-h5n8 (2021).
Wardeh, M., Baylis, M. & Blagrove, M. S. Predicting mammalian hosts in which novel coronaviruses can be generated. Nat. Commun. 12, 780 (2021).
Crossman, L. C. Leveraging deep learning to simulate coronavirus spike proteins has the potential to predict future zoonotic sequences. Preprint at bioRxiv https://doi.org/10.1101/2020.04.20.046920 (2020).
Acknowledgements
The Viral Emergence Research Initiative (VERENA) consortium is supported by NSF BII 2021909. For more information, see viralemergence.org.
Author information
Authors and Affiliations
Contributions
C.J.C. and G.F.A. conceived the study and drafted the manuscript, G.F.A. and C.J.C. produced visualizations, and all authors contributed to the writing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Microbiology thanks Jonathan Dushoff, Vincent Munster and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Albery, G.F., Becker, D.J., Brierley, L. et al. The science of the host–virus network. Nat Microbiol 6, 1483–1492 (2021). https://doi.org/10.1038/s41564-021-00999-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-021-00999-5
This article is cited by
-
Pathogens and planetary change
Nature Reviews Biodiversity (2025)
-
The dual threat: exploring the emergence of human metapneumovirus and SARS-CoV-2 coinfections in respiratory infections
3 Biotech (2025)
-
Identification and genetic characterization of five novel bat coronaviruses from Yunnan, China
BMC Veterinary Research (2024)
-
Predicting host range expansion in parasitic mites using a global mammalian-acarine dataset
Nature Communications (2024)
-
Bacteriophages assisted bacteria to facilitate soil multifunctionality under organochlorine pesticide contamination
Science China Technological Sciences (2024)