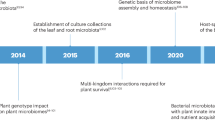
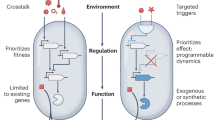
Abstract
The design and use of synthetic communities, or SynComs, is one of the most promising strategies for disentangling the complex interactions within microbial communities, and between these communities and their hosts. Compared to natural communities, these simplified consortia provide the opportunity to study ecological interactions at tractable scales, as well as facilitating reproducibility and fostering interdisciplinary science. However, the effective implementation of the SynCom approach requires several important considerations regarding the development and application of these model systems. There are also emerging ethical considerations when both designing and deploying SynComs in clinical, agricultural or environmental settings. Here we outline current best practices in developing, implementing and evaluating SynComs across different systems, including a focus on important ethical considerations for SynCom research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
de Souza, R. S. C., Armanhi, J. S. L. & Arruda, P. From microbiome to traits: designing synthetic microbial communities for improved crop resiliency. Front. Plant Sci. 11, 1179 (2020).
Schaedler, R. W., Dubs, R. & Costello, R. Association of germfree mice with bacteria isolated from normal mice. J. Exp. Med. 122, 77–82 (1965).
Kim, H. J., Boedicker, J. Q., Choi, J. W. & Ismagilov, R. F. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl Acad. Sci. USA 105, 18188–18193 (2008).
van Leeuwen, P. T., Brul, S., Zhang, J. & Wortel, M. T. Synthetic microbial communities (SynComs) of the human gut: design, assembly and applications. FEMS Microbiol. Rev. 47, fuad012 (2023).
Vorholt, J. A., Vogel, C., Carlström, C. I. & Müller, D. B. Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155 (2017).
Lebeis, S. L. et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864 (2015).
Thonar, C., Frossard, E., Šmilauer, P. & Jansa, J. Competition and facilitation in synthetic communities of arbuscular mycorrhizal fungi. Mol. Ecol. 23, 733–746 (2014).
Durán, P. et al. Microbial interkingdom interactions in roots promote Arabidopsis survival. Cell 175, 973–983.e14 (2018).
Mehlferber, E. C. et al. Phyllosphere microbial associations improve plant reproductive success. Front. Plant Sci. 14, 1273330 (2023).
Flores-Núñez, V. M. et al. Synthetic communities increase microbial diversity and productivity of Agave tequilana plants in the field. Phytobiomes J. 7, 435–448 (2023).
Johns, N. I., Blazejewski, T., Gomes, A. L. & Wang, H. H. Principles for designing synthetic microbial communities. Curr. Opin. Microbiol. 31, 146–153 (2016).
McCarty, N. S. & Ledesma-Amaro, R. Synthetic biology tools to engineer microbial communities for biotechnology. Trends Biotechnol. 37, 181–197 (2019).
Sasse, J. et al. Multilab EcoFAB study shows highly reproducible physiology and depletion of soil metabolites by a model grass. N. Phytologist 222, 1149–1160 (2019).
Theriot, C. M. & Young, V. B. Interactions between the gastrointestinal microbiome and Clostridium difficile. Annu. Rev. Microbiol. 69, 445–461 (2015).
Singer-Englar, T., Barlow, G. & Mathur, R. Obesity, diabetes and the gut microbiome: an updated review. Expert Rev. Gastroenterol. Hepatol. 13, 3–15 (2019).
Vázquez-Castellanos, J. F., Biclot, A., Vrancken, G., Huys, G. R. & Raes, J. Design of synthetic microbial consortia for gut microbiota modulation. Curr. Opin. Pharmacol. 49, 52–59 (2019).
Marín, O., González, B. & Poupin, M. J. From microbial dynamics to functionality in the rhizosphere: a systematic review of the opportunities with synthetic microbial communities. Front. Plant Sci. 12, 650609 (2021).
Peterson, S. B., Bertolli, S. K. & Mougous, J. D. Interbacterial antagonism: at the center of bacterial life. Curr. Biol. 30, R1203–R1214 (2020).
D’Souza, G. et al. Ecology and evolution of metabolic cross-feeding interactions in bacteria. Nat. Prod. Rep. 35, 455–488 (2018).
Pérez Escriva, P., Fuhrer, T. & Sauer, U. Distinct N and C cross-feeding networks in a synthetic mouse gut consortium. mSystems 7, e01484-21 (2022).
Chang, C.-Y., Bajić, D., Vila, J. C. C., Estrela, S. & Sanchez, A. Emergent coexistence in multispecies microbial communities. Science 381, 343–348 (2023).
Finkel, O. M. et al. A single bacterial genus maintains root growth in a complex microbiome. Nature 587, 103–108 (2020).
Anand, G., Goel, V., Dubey, S. & Sharma, S. Tailoring the rhizospheric microbiome of Vigna radiata by adaptation to salt stress. Plant Growth Regul. 93, 79–88 (2021).
Auchtung, J. M., Preisner, E. C., Collins, J., Lerma, A. I. & Britton, R. A. Identification of simplified microbial communities that inhibit Clostridioides difficile infection through dilution/extinction. mSphere 5, e00387-20 (2020).
Kumar, N., Hitch, T. C. A., Haller, D., Lagkouvardos, I. & Clavel, T. MiMiC: a bioinformatic approach for generation of synthetic communities from metagenomes. Microb. Biotechnol. 14, 1757–1770 (2021).
Caballero-Flores, G., Pickard, J. M. & Núñez, G. Microbiota-mediated colonization resistance: mechanisms and regulation. Nat. Rev. Microbiol. 21, 347–360 (2023).
Banerjee, S., Schlaeppi, K. & van der Heijden, M. G. A. Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576 (2018).
Emmenegger, B. et al. Identifying microbiota community patterns important for plant protection using synthetic communities and machine learning. Nat. Commun. 14, 7983 (2023).
Gerna, D., Clara, D., Allwardt, D., Mitter, B. & Roach, T. Tailored media are key to unlocking the diversity of endophytic bacteria in distinct compartments of germinating seeds. Microbiol. Spectr. 10, e00172-22 (2022).
Abisado, R. G., Benomar, S., Klaus, J. R., Dandekar, A. A. & Chandler, J. R. Bacterial quorum sensing and microbial community interaction. mBio 9, e02331-17 (2018).
Rocca, J. D., Muscarella, M. E., Peralta, A. L., Izabel-Shen, D. & Simonin, M. Guided by microbes: applying community coalescence principles for predictive microbiome engineering. mSystems 6, e0053821 (2021).
Debray, R. et al. Priority effects in microbiome assembly. Nat. Rev. Microbiol. 20, 109–121 (2022).
Mutlu, A., Kaspar, C., Becker, N. & Bischofs, I. B. A spore quality-quantity tradeoff favors diverse sporulation strategies in Bacillus subtilis. ISME J. 14, 2703–2714 (2020).
Parnell, J. J., Vintila, S., Tang, C., Wagner, M. R. & Kleiner, M. Evaluation of ready-to-use freezer stocks of a synthetic microbial community for maize root colonization. Microbiol. Spectr. 12, e02401-23 (2023).
Yang, T. et al. Resource availability modulates biodiversity-invasion relationships by altering competitive interactions: resource availability modulates biodiversity. Environ. Microbiol. 19, 2984–2991 (2017).
Maier, B. A. et al. A general non-self response as part of plant immunity. Nat. Plants 7, 696–705 (2021).
Ordon, J. et al. Chromosomal barcodes for simultaneous tracking of near-isogenic bacterial strains in plant microbiota. Nat. Microbiol. 9, 1117–1129 (2024).
Jian, C., Luukkonen, P., Yki-Järvinen, H., Salonen, A. & Korpela, K. Quantitative PCR provides a simple and accessible method for quantitative microbiota profiling. PLoS ONE 15, e0227285 (2020).
Zemb, O. et al. Absolute quantitation of microbes using 16S rRNA gene metabarcoding: a rapid normalization of relative abundances by quantitative PCR targeting a 16S rRNA gene spike-in standard. MicrobiologyOpen 9, e977 (2020).
Morella, N. M., Yang, S. C., Hernandez, C. A. & Koskella, B. Rapid quantification of bacteriophages and their bacterial hosts in vitro and in vivo using droplet digital PCR. J. Virol. Methods 259, 18–24 (2018).
Kembel, S. W., Wu, M., Eisen, J. A. & Green, J. L. Incorporating 16S gene copy number information improves estimates of microbial diversity and abundance. PLoS Comput. Biol. 8, e1002743 (2012).
Brooks, J. P. et al. The truth about metagenomics: quantifying and counteracting bias in 16S rRNA studies. BMC Microbiol. 15, 66 (2015).
Callahan, B. J. et al. DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016).
Callahan, B. J. et al. High-throughput amplicon sequencing of the full-length 16S rRNA gene with single-nucleotide resolution. Nucleic Acids Res. 47, e103 (2019).
Carini, P. et al. Effects of spatial variability and relic DNA removal on the detection of temporal dynamics in soil microbial communities. mBio 11, e02776-19 (2020).
de Souza, R. S. C. et al. Genome sequences of a plant beneficial synthetic bacterial community reveal genetic features for successful plant colonization. Front. Microbiol. 10, 1779 (2019).
Giannoukos, G. et al. Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol. 13, r23 (2012).
Westermann, A. J., Gorski, S. A. & Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 10, 618–630 (2012).
Lewin, G. R. et al. Application of a quantitative framework to improve the accuracy of a bacterial infection model. Proc. Natl Acad. Sci. USA 120, e2221542120 (2023).
Mohajeri, M. H. et al. The role of the microbiome for human health: from basic science to clinical applications. Eur. J. Nutr. 57, 1–14 (2018).
Mallott, E. K. et al. Human microbiome variation associated with race and ethnicity emerges as early as 3 months of age. PLoS Biol. 21, e3002230 (2023).
Gupta, V. K., Paul, S. & Dutta, C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front. Microbiol. 8, 1162 (2017).
DeFilipp, Z. et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019).
Varga, J. J. et al. Antibiotics drive expansion of rare pathogens in a chronic infection microbiome model. mSphere 7, e00318-22 (2022).
Bogaert, D. et al. Mother-to-infant microbiota transmission and infant microbiota development across multiple body sites. Cell Host Microbe 31, 447–460.e6 (2023).
Ladau, J. et al. Microbial invasions and inoculants: a call to action. Preprint at https://ecoevorxiv.org/repository/view/5702/ (2023).
Lange, L. et al. Microbiome ethics, guiding principles for microbiome research, use and knowledge management. Environ. Microbiome 17, 50 (2022).
Pantoja Angles, A., Valle-Pérez, A. U., Hauser, C. & Mahfouz, M. M. Microbial biocontainment systems for clinical, agricultural and industrial applications. Front. Bioeng. Biotechnol. 10, 830200 (2022).
Huttenhower, C., Finn, R. D. & McHardy, A. C. Challenges and opportunities in sharing microbiome data and analyses. Nat. Microbiol. 8, 1960–1970 (2023).
Yilmaz, P. et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nat. Biotechnol. 29, 415–420 (2011).
Mirzayi, C. et al. Reporting guidelines for human microbiome research: the STORMS checklist. Nat. Med. 27, 1885–1892 (2021).
Northen, T. R. et al. Community standards and future opportunities for synthetic communities in plant–microbiota research. Nat. Microbiol. https://doi.org/10.1038/s41564-024-01833-4 (2024).
Jennings, S. A. V. & Clavel, T. Synthetic communities of gut microbes for basic research and translational approaches in animal health and nutrition. Annu. Rev. Anim. Biosci. 12, 283–300 (2024).
Shayanthan, A., Ordoñez, P. A. C. & Oresnik, I. J. The role of synthetic microbial communities (SynCom) in sustainable agriculture. Front. Agron 4, 896307 (2022).
Cheng, A. G. et al. Design, construction and in vivo augmentation of a complex gut microbiome. Cell 185, 3617–3636.e19 (2022).
Schloss, P. D. et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541 (2009).
Edgar, R. C. UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. Preprint at https://doi.org/10.1101/081257 (2016).
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C. & Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200 (2011).
Davis, N. M., Proctor, D. M., Holmes, S. P., Relman, D. A. & Callahan, B. J. Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018).
Frøslev, T. G. et al. Algorithm for post-clustering curation of DNA amplicon data yields reliable biodiversity estimates. Nat. Commun. 8, 1188 (2017).
Dixon, P. VEGAN, a package of R functions for community ecology. J. Vegetation Sci. 14, 927–930 (2003).
McMurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Barnett, D. J. M., Arts, I. C. W. & Penders, J. microViz: an R package for microbiome data visualization and statistics. J. Open Source Softw. 6, 3201 (2021).
Lahti, L. & Shetty, S. microbiome R package https://doi.org/10.18129/B9.bioc.microbiome (Bioconductor, 2017).
Bankevich, A. et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477 (2012).
Wick, R. R., Judd, L. M., Gorrie, C. L. & Holt, K. E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13, e1005595 (2017).
Schwengers, O. et al. Bakta: rapid and standardized annotation of bacterial genomes via alignment-free sequence identification. Micro. Genom. 7, 000685 (2021).
Shade, A. & Stopnisek, N. Abundance-occupancy distributions to prioritize plant core microbiome membership. Curr. Opin. Microbiol. 49, 50–58 (2019).
Fisher, C. K. & Mehta, P. Identifying keystone species in the human gut microbiome from metagenomic timeseries using sparse linear regression. PLoS ONE 9, e102451 (2014).
Wang, X.-W. et al. Identifying keystone species in microbial communities using deep learning. Nat. Ecol. Evol. 8, 22–31 (2024).
Kurtz, Z. D. et al. Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11, e1004226 (2015).
Karkaria, B. D., Fedorec, A. J. H. & Barnes, C. P. Automated design of synthetic microbial communities. Nat. Commun. 12, 672 (2021).
Toju, H. et al. Scoring species for synthetic community design: network analyses of functional core microbiomes. Front. Microbiol. 11, 1361 (2020).
Paredes, S. H. et al. Design of synthetic bacterial communities for predictable plant phenotypes. PLoS Biol. 16, e2003962 (2018).
Acknowledgements
This manuscript was inspired by an ASM Microbe panel discussion involving many of the authors and organized by the ASM EEB track in 2023. We would like to specifically thank B. Callahan for early discussions on this topic. E.C.M. acknowledges funding from the NSF EAGER award no. 1838299, as well as support from the NSF Postdoctoral Research Fellowships in Biology award no. 2209151. B.K. is a Chan Zuckerberg San Francisco Biohub investigator. B.J. and K.A.P. acknowledge funding from the National Institutes of Health award no. U19 AI157981. G.A. and M.S. were supported by the 3rd Programme for Future Investments (France 2030), operated by the SUCSEED project (ANR- 20- PCPA-0009) and funded by the ‘Growing and Protecting crops Differently’ French Priority Research Program (PPR-CPA), part of the national investment plan operated by the French National Research Agency (ANR). L.P.P.-M. acknowledges funding from Conahcyt (Consejo Nacional de Humanidades, Ciencias y Tecnologías) under awards nos. A1-S-9889 and CBF2023-2024-2642.
Author information
Authors and Affiliations
Contributions
E.C.M. and B.K. developed the framework for the manuscript, with input from all authors. E.C.M. led the writing of the manuscript, with contributions by G.A., B.J., L.P.P.-M., K.A.P., M.S. and B.K. Authors G.A., B.J., L.P.P.-M. and K.A.P. contributed equally to the manuscript; these authors are presented in alphabetical order by surname.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mehlferber, E.C., Arnault, G., Joshi, B. et al. A cross-systems primer for synthetic microbial communities. Nat Microbiol 9, 2765–2773 (2024). https://doi.org/10.1038/s41564-024-01827-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41564-024-01827-2
This article is cited by
-
Phyllosphere synthetic microbial communities: a new frontier in plant protection
BMC Plant Biology (2025)
-
Synthetic microbial consortia based on quorum-sensing for disease therapy
Bioresources and Bioprocessing (2025)
-
Building microbial communities to improve antimicrobial strategies
npj Antimicrobials and Resistance (2025)
-
Community standards and future opportunities for synthetic communities in plant–microbiota research
Nature Microbiology (2024)