Abstract
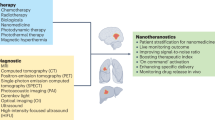
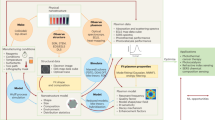
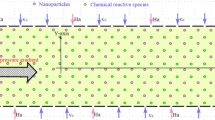
The inherent limits of traditional diagnoses and therapies have driven the development and application of emerging nanotechnologies for more effective and safer management of diseases, herein referred to as ‘nanotheranostics’. Although many important technological successes have been achieved in this field, widespread adoption of nanotheranostics as a new paradigm is hindered by specific obstacles, including time-consuming synthesis of nanoparticles, incomplete understanding of nano–bio interactions, and challenges regarding chemistry, manufacturing and the controls required for clinical translation and commercialization. As a key branch of artificial intelligence, machine learning (ML) provides a set of tools capable of performing time-consuming and result-perception tasks, thus offering unique opportunities for nanotheranostics. This Review summarizes the progress and challenges in this emerging field of ML-aided nanotheranostics, and discusses the opportunities in developing next-generation nanotheranostics with reliable datasets and advanced ML models to offer better clinical benefits to patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Chen, H., Zhang, W., Zhu, G., Xie, J. & Chen, X. Rethinking cancer nanotheranostics. Nat. Rev. Mater. 2, 17024 (2017).
Shi, J., Kantoff, P. W., Wooster, R. & Farokhzad, O. C. Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer 17, 20–37 (2017).
AbdElFatah, T. et al. Nanoplasmonic amplification in microfluidics enables accelerated colorimetric quantification of nucleic acid biomarkers from pathogens. Nat. Nanotechnol. 18, 922–932 (2023).
Mitchell, M. J. et al. Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov. 20, 101–124 (2021).
Hou, X., Zaks, T., Langer, R. & Dong, Y. Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater. 6, 1078–1094 (2021).
Kim, M. et al. Detection of ovarian cancer via the spectral fingerprinting of quantum-defect-modified carbon nanotubes in serum by machine learning. Nat. Biomed. Eng. 6, 267–275 (2022).
Zhang, J., Zhao, T., Jakobsson, V. & Chen, X. Clinical translation of radiotheranostics for precision oncology. Nat. Rev. Bioeng. 1, 612–614 (2023).
Fang, R. H., Gao, W. & Zhang, L. Targeting drugs to tumours using cell membrane-coated nanoparticles. Nat. Rev. Clin. Oncol. 20, 33–48 (2023).
Li, X., Lovell, J. F., Yoon, J. & Chen, X. Clinical development and potential of photothermal and photodynamic therapies for cancer. Nat. Rev. Clin. Oncol. 17, 657–674 (2020).
Raguram, A., Banskota, S. & Liu, D. R. Therapeutic in vivo delivery of gene editing agents. Cell 185, 2806–2827 (2022).
Nam, J. et al. Cancer nanomedicine for combination cancer immunotherapy. Nat. Rev. Mater. 4, 398–414 (2019).
Zhao, H. et al. A robotic platform for the synthesis of colloidal nanocrystals. Nat. Synth. 2, 505–514 (2023).
Huang, X. et al. Nanotechnology-based strategies against SARS-CoV-2 variants. Nat. Nanotechnol. 17, 1027–1037 (2022).
Park, J. et al. An integrated magneto-electrochemical device for the rapid profiling of tumour extracellular vesicles from blood plasma. Nat. Biomed. Eng. 5, 678–689 (2021).
Rao, L. et al. Hybrid cellular membrane nanovesicles amplify macrophage immune responses against cancer recurrence and metastasis. Nat. Commun. 11, 4909 (2020).
Butler, K. T., Davies, D. W., Cartwright, H., Isayev, O. & Walsh, A. Machine learning for molecular and materials science. Nature 559, 547–555 (2018).
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Shen, D., Wu, G. & Suk, H.-I. Deep learning in medical image analysis. Annu. Rev. Biomed. Eng. 19, 221–248 (2017).
Camacho, D. M., Collins, K. M., Powers, R. K., Costello, J. C. & Collins, J. J. Next-generation machine learning for biological networks. Cell 173, 1581–1592 (2018).
Yu, K.-H., Beam, A. L. & Kohane, I. S. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2, 719–731 (2018).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Segler, M. H. S., Preuss, M. & Waller, M. P. Planning chemical syntheses with deep neural networks and symbolic AI. Nature 555, 604–610 (2018).
Ziatdinov, M., Ghosh, A., Wong, C. Y. & Kalinin, S. V. AtomAI framework for deep learning analysis of image and spectroscopy data in electron and scanning probe microscopy. Nat. Mach. Intell. 4, 1101–1112 (2022).
Heinzmann, K., Carter, L. M., Lewis, J. S. & Aboagye, E. O. Multiplexed imaging for diagnosis and therapy. Nat. Biomed. Eng. 1, 697–713 (2017).
Acosta, J. N., Falcone, G. J., Rajpurkar, P. & Topol, E. J. Multimodal biomedical AI. Nat. Med. 28, 1773–1784 (2022).
Wong, F., de la Fuente-Nunez, C. & Collins, J. J. Leveraging artificial intelligence in the fight against infectious diseases. Science 381, 164–170 (2023).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Chih-Wei, H. & Chih-Jen, L. A comparison of methods for multiclass support vector machines. IEEE Trans. Neural Netw. 13, 415–425 (2002).
Wold, S., Sjöström, M. & Eriksson, L. PLS-regression: a basic tool of chemometrics. Chemometr. Intell. Lab. Syst. 58, 109–130 (2001).
Masson, J.-F., Biggins, J. S. & Ringe, E. Machine learning for nanoplasmonics. Nat. Nanotechnol. 18, 111–123 (2023).
Wan, F., Wong, F., Collins, J. J. & de la Fuente-Nunez, C. Machine learning for antimicrobial peptide identification and design. Nat. Rev. Bioeng. 2, 392–407 (2024).
Mahmoudi, M., Landry, M. P., Moore, A. & Coreas, R. The protein corona from nanomedicine to environmental science. Nat. Rev. Mater. 8, 422–438 (2023).
Tao, H. et al. Nanoparticle synthesis assisted by machine learning. Nat. Rev. Mater. 6, 701–716 (2021).
Dai, X. & Chen, Y. Computational biomaterials: computational simulations for biomedicine. Adv. Mater. 35, 2204798 (2023).
Merchant, A. et al. Scaling deep learning for materials discovery. Nature 624, 80–85 (2023).
Batra, R. et al. Machine learning overcomes human bias in the discovery of self-assembling peptides. Nat. Chem. 14, 1427–1435 (2022).
Zhu, M. et al. Machine-learning-assisted single-vessel analysis of nanoparticle permeability in tumour vasculatures. Nat. Nanotechnol. 18, 657–666 (2023).
Boehnke, N. et al. Massively parallel pooled screening reveals genomic determinants of nanoparticle delivery. Science 377, eabm5551 (2022).
Yamankurt, G. et al. Exploration of the nanomedicine-design space with high-throughput screening and machine learning. Nat. Biomed. Eng. 3, 318–327 (2019).
Shamay, Y. et al. Quantitative self-assembly prediction yields targeted nanomedicines. Nat. Mater. 17, 361–368 (2018).
Stater, E. P., Sonay, A. Y., Hart, C. & Grimm, J. The ancillary effects of nanoparticles and their implications for nanomedicine. Nat. Nanotechnol. 16, 1180–1194 (2021).
Lu, Y., Aimetti, A. A., Langer, R. & Gu, Z. Bioresponsive materials. Nat. Rev. Mater. 1, 16075 (2016).
Hong, G., Diao, S., Antaris, A. L. & Dai, H. Carbon nanomaterials for biological imaging and nanomedicinal therapy. Chem. Rev. 115, 10816–10906 (2015).
Suwardi, A. et al. Machine learning-driven biomaterials evolution. Adv. Mater. 34, 2102703 (2022).
Rycenga, M. et al. Controlling the synthesis and assembly of silver nanostructures for plasmonic applications. Chem. Rev. 111, 3669–3712 (2011).
Yang, X., Yang, M., Pang, B., Vara, M. & Xia, Y. Gold nanomaterials at work in biomedicine. Chem. Rev. 115, 10410–10488 (2015).
Michalet, X. et al. Quantum dots for live cells, in vivo imaging, and diagnostics. Science 307, 538–544 (2005).
Kim, P. et al. Quantifying the efficacy of magnetic nanoparticles for MRI and hyperthermia applications via machine learning methods. Small 19, 2303522 (2023).
Serov, N. & Vinogradov, V. Artificial intelligence to bring nanomedicine to life. Adv. Drug Deliv. Rev. 184, 114194 (2022).
Grand, J., Auguié, B. & Le Ru, E. C. Combined extinction and absorption UV–visible spectroscopy as a method for revealing shape imperfections of metallic nanoparticles. Anal. Chem. 91, 14639–14648 (2019).
Gherman, A. M. M. et al. Artificial neural networks modeling of the parameterized gold nanoparticles generation through photo-induced process. Mater. Res. Express 5, 085011 (2018).
Shafaei, A. & Khayati, G. R. A predictive model on size of silver nanoparticles prepared by green synthesis method using hybrid artificial neural network–particle swarm optimization algorithm. Measurement 151, 107199 (2020).
Orimoto, Y. et al. Application of artificial neural networks to rapid data analysis in combinatorial nanoparticle syntheses. J. Phys. Chem. C 116, 17885–17896 (2012).
Salley, D. et al. A nanomaterials discovery robot for the Darwinian evolution of shape programmable gold nanoparticles. Nat. Commun. 11, 2771 (2020).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue-specific mRNA delivery and CRISPR–Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Ng, K. K. & Zheng, G. Molecular interactions in organic nanoparticles for phototheranostic applications. Chem. Rev. 115, 11012–11042 (2015).
Andrews, N. et al. COVID-19 vaccine effectiveness against the Omicron (B.1.1.529) variant. N. Engl. J. Med. 386, 1532–1546 (2022).
Li, B. et al. Combinatorial design of nanoparticles for pulmonary mRNA delivery and genome editing. Nat. Biotechnol. 41, 1410–1415 (2023).
Wang, W. et al. Prediction of lipid nanoparticles for mRNA vaccines by the machine learning algorithm. Acta Pharm. Sin. B 12, 2950–2962 (2022).
Walkey, C. D. & Chan, W. C. W. Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment. Chem. Soc. Rev. 41, 2780–2799 (2012).
Youshia, J., Ali, M. E. & Lamprecht, A. Artificial neural network based particle size prediction of polymeric nanoparticles. Eur. J. Pharm. Biopharm. 119, 333–342 (2017).
Shalaby, K. S. et al. Determination of factors controlling the particle size and entrapment efficiency of noscapine in PEG/PLA nanoparticles using artificial neural networks. Int. J. Nanomed. 9, 4953–4964 (2014).
Ogden, P. J., Kelsic, E. D., Sinai, S. & Church, G. M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science 366, 1139–1143 (2019).
Meng, Q.-F. et al. Inhalation delivery of dexamethasone with iSEND nanoparticles attenuates the COVID-19 cytokine storm in mice and nonhuman primates. Sci. Adv. 9, eadg3277 (2023).
Wilhelm, S. et al. Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater. 1, 16014 (2016).
Herrmann, I. K., Wood, M. J. A. & Fuhrmann, G. Extracellular vesicles as a next-generation drug delivery platform. Nat. Nanotechnol. 16, 748–759 (2021).
Madigan, V., Zhang, F. & Dahlman, J. E. Drug delivery systems for CRISPR-based genome editors. Nat. Rev. Drug Discov. 22, 875–894 (2023).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Zengel, J. et al. Hardwiring tissue-specific AAV transduction in mice through engineered receptor expression. Nat. Methods 20, 1070–1081 (2023).
Bryant, D. H. et al. Deep diversification of an AAV capsid protein by machine learning. Nat. Biotechnol. 39, 691–696 (2021).
El Andaloussi, S., Mäger, I., Breakefield, X. O. & Wood, M. J. A. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013).
Zheng, W. et al. Diagnosis of paediatric tuberculosis by optically detecting two virulence factors on extracellular vesicles in blood samples. Nat. Biomed. Eng. 6, 979–991 (2022).
Kuypers, S. et al. Unsupervised machine learning-based clustering of nanosized fluorescent extracellular vesicles. Small 17, 2006786 (2021).
Mahmoudi, M. et al. Protein−nanoparticle interactions: opportunities and challenges. Chem. Rev. 111, 5610–5637 (2011).
Salvati, A. et al. Transferrin-functionalized nanoparticles lose their targeting capabilities when a biomolecule corona adsorbs on the surface. Nat. Nanotechnol. 8, 137–143 (2013).
Nel, A. E. et al. Understanding biophysicochemical interactions at the nano–bio interface. Nat. Mater. 8, 543–557 (2009).
Kingston, B. R., Syed, A. M., Ngai, J., Sindhwani, S. & Chan, W. C. W. Assessing micrometastases as a target for nanoparticles using 3D microscopy and machine learning. Proc. Natl Acad. Sci. USA 116, 14937–14946 (2019).
Ferdosi, S. et al. Engineered nanoparticles enable deep proteomics studies at scale by leveraging tunable nano–bio interactions. Proc. Natl Acad. Sci. USA 119, e2106053119 (2022).
Cha, M. et al. Unifying structural descriptors for biological and bioinspired nanoscale complexes. Nat. Comput. Sci. 2, 243–252 (2022).
Ban, Z. et al. Machine learning predicts the functional composition of the protein corona and the cellular recognition of nanoparticles. Proc. Natl Acad. Sci. USA 117, 10492–10499 (2020).
Ouassil, N., Pinals, R. L., Del Bonis-O’Donnell, J. T., Wang, J. W. & Landry, M. P. Supervised learning model predicts protein adsorption to carbon nanotubes. Sci. Adv. 8, eabm0898 (2022).
Saldinger, J. C., Raymond, M., Elvati, P. & Violi, A. Domain-agnostic predictions of nanoscale interactions in proteins and nanoparticles. Nat. Comput. Sci. 3, 393–402 (2023).
Liu, R., Jiang, W., Walkey, C. D., Chan, W. C. W. & Cohen, Y. Prediction of nanoparticles–cell association based on corona proteins and physicochemical properties. Nanoscale 7, 9664–9675 (2015).
Lazarovits, J. et al. Supervised learning and mass spectrometry predicts the in vivo fate of nanomaterials. ACS Nano 13, 8023–8034 (2019).
Fourches, D. et al. Quantitative nanostructure−activity relationship modeling. ACS Nano 4, 5703–5712 (2010).
Behzadi, S. et al. Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev. 46, 4218–4244 (2017).
Walkey, C. D. et al. Protein corona fingerprinting predicts the cellular interaction of gold and silver nanoparticles. ACS Nano 8, 2439–2455 (2014).
Loecher, A., Bruyns-Haylett, M., Ballester, P. J., Borros, S. & Oliva, N. A machine learning approach to predict cellular uptake of pBAE polyplexes. Biomater. Sci. 11, 5797–5808 (2023).
Shirokii, N. et al. Quantitative prediction of inorganic nanomaterial cellular toxicity via machine learning. Small 19, 2207106 (2023).
Martin et al. Evidence-based prediction of cellular toxicity for amorphous silica nanoparticles. ACS Nano 17, 9987–9999 (2023).
Jyakhwo, S., Serov, N., Dmitrenko, A. & Vinogradov, V. V. Machine learning reinforced genetic algorithm for massive targeted discovery of selectively cytotoxic inorganic nanoparticles. Small 20, 2305375 (2024).
Puzyn, T. et al. Using nano-QSAR to predict the cytotoxicity of metal oxide nanoparticles. Nat. Nanotechnol. 6, 175–178 (2011).
Sealfon, R. S. G., Wong, A. K. & Troyanskaya, O. G. Machine learning methods to model multicellular complexity and tissue specificity. Nat. Rev. Mater. 6, 717–729 (2021).
Chen, Q. et al. Meta-analysis of nanoparticle distribution in tumors and major organs in tumor-bearing mice. ACS Nano 17, 19810–19831 (2023).
MacMillan, P. et al. Toward predicting nanoparticle distribution in heterogeneous tumor tissues. Nano Lett. 23, 7197–7205 (2023).
Liu, X. et al. Predictive modeling of nanomaterial exposure effects in biological systems. Int. J. Nanomed. 8, 31–43 (2023).
Gilbertson, L. M. et al. Toward safer multi-walled carbon nanotube design: establishing a statistical model that relates surface charge and embryonic zebrafish mortality. Nanotoxicology 10, 10–19 (2016).
Song, Y. et al. 3D-printed epifluidic electronic skin for machine learning-powered multimodal health surveillance. Sci. Adv. 9, eadi6492 (2023).
Lin, A. A., Nimgaonkar, V., Issadore, D. & Carpenter, E. L. Extracellular vesicle-based multianalyte liquid biopsy as a diagnostic for cancer. Annu. Rev. Biomed. Data Sci. 5, 269–292 (2022).
Xu, C., Solomon, S. A. & Gao, W. Artificial intelligence-powered electronic skin. Nat. Mach. Intell. 5, 1344–1355 (2023).
Altug, H., Oh, S.-H., Maier, S. A. & Homola, J. Advances and applications of nanophotonic biosensors. Nat. Nanotechnol. 17, 5–16 (2022).
Safir, F. et al. Combining acoustic bioprinting with AI-assisted raman spectroscopy for high-throughput identification of bacteria in blood. Nano Lett. 23, 2065–2073 (2023).
Shin, H. et al. Single test-based diagnosis of multiple cancer types using exosome-SERS-AI for early stage cancers. Nat. Commun. 14, 1644 (2023).
Kavungal, D. et al. Artificial intelligence-coupled plasmonic infrared sensor for detection of structural protein biomarkers in neurodegenerative diseases. Sci. Adv. 9, eadg9644 (2023).
Gao, Z. et al. Machine-learning-assisted microfluidic nanoplasmonic digital immunoassay for cytokine storm profiling in COVID-19 patients. ACS Nano 15, 18023–18036 (2021).
Thrift, W. J. et al. Deep learning analysis of vibrational spectra of bacterial lysate for rapid antimicrobial susceptibility testing. ACS Nano 14, 15336–15348 (2020).
Wang, Y., Zhao, Y., Bollas, A., Wang, Y. & Au, K. F. Nanopore sequencing technology, bioinformatics and applications. Nat. Biotechnol. 39, 1348–1365 (2021).
Zhang, M. et al. Real-time detection of 20 amino acids and discrimination of pathologically relevant peptides with functionalized nanopore. Nat. Methods 21, 609–618 (2024).
Ying, Y.-L. et al. Nanopore-based technologies beyond DNA sequencing. Nat. Nanotechnol. 17, 1136–1146 (2022).
Jena, M. K. & Pathak, B. Development of an artificially intelligent nanopore for high-throughput DNA sequencing with a machine-learning-aided quantum-tunneling approach. Nano Lett. 23, 2511–2521 (2023).
Taniguchi, M. et al. Combining machine learning and nanopore construction creates an artificial intelligence nanopore for coronavirus detection. Nat. Commun. 12, 3726 (2021).
Xia, K. et al. Synthetic heparan sulfate standards and machine learning facilitate the development of solid-state nanopore analysis. Proc. Natl Acad. Sci. USA 118, e2022806118 (2021).
Li, M. et al. Identification of tagged glycans with a protein nanopore. Nat. Commun. 14, 1737 (2023).
Wang, Y. et al. Identification of nucleoside monophosphates and their epigenetic modifications using an engineered nanopore. Nat. Nanotechnol. 17, 976–983 (2022).
Greive, S. J., Bacri, L., Cressiot, B. & Pelta, J. Identification of conformational variants for bradykinin biomarker peptides from a biofluid using a nanopore and machine learning. ACS Nano 18, 539–550 (2024).
Sajda, P. Machine learning for detection and diagnosis of disease. Annu. Rev. Biomed. Eng. 8, 537–565 (2006).
Tian, F. et al. Protein analysis of extracellular vesicles to monitor and predict therapeutic response in metastatic breast cancer. Nat. Commun. 12, 2536 (2021).
Sahu, A. et al. Regulation of aged skeletal muscle regeneration by circulating extracellular vesicles. Nat. Aging 1, 1148–1161 (2021).
Mangalwedhekar, R. et al. Achieving nanoscale precision using neuromorphic localization microscopy. Nat. Nanotechnol. 18, 380–389 (2023).
Reis, M. et al. Machine-learning-guided discovery of 19F MRI agents enabled by automated copolymer synthesis. J. Am. Chem. Soc. 143, 17677–17689 (2021).
Ma, Z., Wang, F., Wang, W., Zhong, Y. & Dai, H. Deep learning for in vivo near-infrared imaging. Proc. Natl Acad. Sci. USA 118, e2021446118 (2021).
Moen, E. et al. Deep learning for cellular image analysis. Nat. Methods 16, 1233–1246 (2019).
Bouchard, C. et al. Resolution enhancement with a task-assisted GAN to guide optical nanoscopy image analysis and acquisition. Nat. Mach. Intell. 5, 830–844 (2023).
Park, J. et al. Artificial intelligence-enabled quantitative phase imaging methods for life sciences. Nat. Methods 20, 1645–1660 (2023).
Shmatko, A., Ghaffari Laleh, N., Gerstung, M. & Kather, J. N. Artificial intelligence in histopathology: enhancing cancer research and clinical oncology. Nat. Cancer 3, 1026–1038 (2022).
Hong, G. et al. Through-skull fluorescence imaging of the brain in a new near-infrared window. Nat. Photon. 8, 723–730 (2014).
Chen, X. et al. Artificial confocal microscopy for deep label-free imaging. Nat. Photon. 17, 250–258 (2023).
Ham, D., Park, H., Hwang, S. & Kim, K. Neuromorphic electronics based on copying and pasting the brain. Nat. Electron. 4, 635–644 (2021).
Oumano, M. & Yu, H. A deep learning approach to gold nanoparticle quantification in computed tomography. Phys. Med. 87, 83–89 (2021).
Reker, D. et al. Computationally guided high-throughput design of self-assembling drug nanoparticles. Nat. Nanotechnol. 16, 725–733 (2021).
Hsueh, H. T. et al. Machine learning-driven multifunctional peptide engineering for sustained ocular drug delivery. Nat. Commun. 14, 2509 (2023).
Castillo-Hair, S. M. & Seelig, G. Machine learning for designing next-generation mRNA therapeutics. Acc. Chem. Res. 55, 24–34 (2022).
Zhang, H. et al. Algorithm for optimized mRNA design improves stability and immunogenicity. Nature 621, 396–403 (2023).
Ebrahimi, S. B., Samanta, D., Kusmierz, C. D. & Mirkin, C. A. Protein transfection via spherical nucleic acids. Nat. Protoc. 17, 327–357 (2022).
Huang, J. et al. Identification of potent antimicrobial peptides via a machine-learning pipeline that mines the entire space of peptide sequences. Nat. Biomed. Eng. 7, 797–810 (2023).
O’Callaghan, J. How OpenAI’s text-to-video tool Sora could change science—and society. Nature 627, 475–476 (2024).
Thorp, H. H. ChatGPT is fun, but not an author. Science 379, 313 (2023).
Tropsha, A., Mills, K. C. & Hickey, A. J. Reproducibility, sharing and progress in nanomaterial databases. Nat. Nanotechnol. 12, 1111–1114 (2017).
de la Iglesia, D. et al. A machine learning approach to identify clinical trials involving nanodrugs and nanodevices from ClinicalTrials.gov. PLoS ONE 9, e110331 (2014).
Wyrzykowska, E. et al. Representing and describing nanomaterials in predictive nanoinformatics. Nat. Nanotechnol. 17, 924–932 (2022).
Ekins, S. et al. Exploiting machine learning for end-to-end drug discovery and development. Nat. Mater. 18, 435–441 (2019).
Erion, G. et al. A cost-aware framework for the development of AI models for healthcare applications. Nat. Biomed. Eng. 6, 1384–1398 (2022).
Yan, X., Sedykh, A., Wang, W., Yan, B. & Zhu, H. Construction of a web-based nanomaterial database by big data curation and modeling friendly nanostructure annotations. Nat. Commun. 11, 2519 (2020).
Wang, Y. & Kohane, D. S. External triggering and triggered targeting strategies for drug delivery. Nat. Rev. Mater. 2, 17020 (2017).
Ling, Q., Herstine, J. A., Bradbury, A. & Gray, S. J. AAV-based in vivo gene therapy for neurological disorders. Nat. Rev. Drug Discov. 22, 789–806 (2023).
Hu, S. et al. A mussel-inspired film for adhesion to wet buccal tissue and efficient buccal drug delivery. Nat. Commun. 12, 1689 (2021).
Acknowledgements
This work was supported by National Natural Science Foundation of China (82222035 and 82372106), Shenzhen Medical Research Fund (B2302041), Shenzhen Bay Laboratory Proof-of-Concept Fund (S231801005), National Medical Research Council of Singapore (MOH-001388-00 and CG21APR1005), Singapore Ministry of Education (MOE-000387-00) and National University of Singapore (NUHSRO/2021/034/TRP/09/Nanomedicine).
Author information
Authors and Affiliations
Contributions
L.R. contributed the idea and completed the figures. Y.Y., X.S., G.Y. and X.C. helped to write, review and revise the paper.
Corresponding authors
Ethics declarations
Competing interests
X.S. is the co-founder and CTO of Shenzhen Intellindust. X.C. is the co-founder and CTO of Yantai LNC Biotechnology. The other authors declare no competing interests.
Peer review
Peer review information
Nature Nanotechnology thanks Milad Abolhasani, Liangfang Zhang and Hao Zhu for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Rao, L., Yuan, Y., Shen, X. et al. Designing nanotheranostics with machine learning. Nat. Nanotechnol. 19, 1769–1781 (2024). https://doi.org/10.1038/s41565-024-01753-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41565-024-01753-8
This article is cited by
-
Emerging nano-immunotherapeutic strategies achieve metastatic colorectal cancer precision therapy
Journal of Nanobiotechnology (2026)
-
Harnessing nanomedicine to target NF-κB signalling in cancer: at the intersection of inflammatory signalling, metabolic reprogramming, and therapeutic innovation
Cell Communication and Signaling (2026)
-
Nanoparticle-mediated targeting chimeras transform targeted protein degradation
Nature Nanotechnology (2026)
-
Synergistic Ferroptosis–Immunotherapy Nanoplatforms: Multidimensional Engineering for Tumor Microenvironment Remodeling and Therapeutic Optimization
Nano-Micro Letters (2026)
-
Convergence in Nanomedicine: Integrating Brain-Targeted Delivery and Gut Microbiota Modulation for Neurological Protection
BioNanoScience (2026)