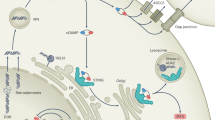
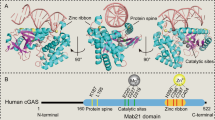
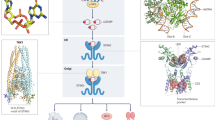
Abstract
The cyclic guanosine monophosphate–adenosine monophosphate synthase (cGAS)–stimulator of interferon genes (STING) pathway has a crucial role in detecting tumour-derived DNA, whether the pathway is generated spontaneously or induced therapeutically. Activation of the cGAS–STING pathway triggers type I interferon signalling and pro-inflammatory responses in both tumour and immune cells, establishing a delicate balance between pathological inflammation and protective immune responses. Although preclinical studies have highlighted the promise of targeting the cGAS–STING pathway to enhance antitumour immunotherapy, clinical results have fallen short of expectations. In this Review, we outline key advances in understanding the tumour-promoting and tumour-suppressive effects mediated by the cGAS–STING pathway and discuss opportunities and challenges for its integration into future cancer immunotherapy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Matthews, H. K., Bertoli, C. & de Bruin, R. A. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 23, 74–88 (2022).
Bakhoum, S. F. & Cantley, L. C. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 174, 1347–1360 (2018).
Sun, L., Wu, J., Du, F., Chen, X. & Chen, Z. J. Cyclic GMP–AMP synthase is a cytosolic DNA sensor that activates the type I interferon pathway. Science 339, 786–791 (2013).
Shang, G., Zhang, C., Chen, Z. J., Bai, X. C. & Zhang, X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP–AMP. Nature 567, 389–393 (2019).
Ergun, S. L., Fernandez, D., Weiss, T. M. & Li, L. STING polymer structure reveals mechanisms for activation, hyperactivation, and inhibition. Cell 178, 290–301.e210 (2019).
Dobbs, N. et al. STING activation by translocation from the ER is associated with infection and autoinflammatory disease. Cell Host Microbe 18, 157–168 (2015).
Dong, M. & Fitzgerald, K. A. DNA-sensing pathways in health, autoinflammatory and autoimmune diseases. Nat. Immunol. 25, 2001–2014 (2024).
Zhu, H. et al. Targeting DNA damage response pathways in tumor drug resistance: mechanisms, clinical implications, and future directions. Drug Resist. Updates 83, 101287 (2025).
Barber, G. N. STING: infection, inflammation and cancer. Nat. Rev. Immunol. 15, 760–770 (2015).
Vilenchik, M. M. & Knudson, A. G. Endogenous DNA double-strand breaks: production, fidelity of repair, and induction of cancer. Proc. Natl Acad. Sci. USA 100, 12871–12876 (2003).
Goldstein, M. & Kastan, M. B. The DNA damage response: implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 66, 129–143 (2015).
Konno, H. et al. Pro-inflammation associated with a gain-of-function mutation (R284S) in the innate immune sensor STING. Cell Rep. 23, 1112–1123 (2018).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Drews, R. M. et al. A pan-cancer compendium of chromosomal instability. Nature 606, 976–983 (2022).
Crasta, K. et al. DNA breaks and chromosome pulverization from errors in mitosis. Nature 482, 53–58 (2012).
Mackenzie, K. J. et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 548, 461–465 (2017).
Harding, S. M. et al. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 548, 466–470 (2017).
Bakhoum, S. F. et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 553, 467–472 (2018).
Li, J. et al. Non-cell-autonomous cancer progression from chromosomal instability. Nature 620, 1080–1088 (2023). This study shows that CIN-mediated engagement of the cGAS–STING pathway promotes metastasis in a TME-dependent manner and that pharmacological inhibition of STING suppresses metastasis.
Hong, C. et al. cGAS–STING drives the IL-6-dependent survival of chromosomally instable cancers. Nature 607, 366–373 (2022). This study demonstrates that cancer cells produce the interleukin IL-6 via CIN-driven activation of the cGAS–STING noncanonical NF-κB pathway to promote their own proliferation.
Mitelman, F. Mitelman Database: chromosome aberrations and gene fusions in cancer. Mitelman Database https://mitelmandatabase.isb-cgc.org/ (2025).
Cimini, D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim. Biophys. Acta Rev. Cancer 1786, 32–40 (2008).
Carter, S. L., Eklund, A. C., Kohane, I. S., Harris, L. N. & Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 38, 1043–1048 (2006).
Takaki, T., Millar, R., Hiley, C. T. & Boulton, S. J. Micronuclei induced by radiation, replication stress, or chromosome segregation errors do not activate cGAS–STING. Mol. Cell 84, 2203–2213.e2205 (2024).
Davoli, T., Uno, H., Wooten, E. C. & Elledge, S. J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 355, eaaf8399 (2017).
Agustinus, A. S. et al. Epigenetic dysregulation from chromosomal transit in micronuclei. Nature 619, 176–183 (2023).
Gaillard, H., García-Muse, T. & Aguilera, A. Replication stress and cancer. Nat. Rev. Cancer 15, 276–289 (2015).
Hanada, K. et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 14, 1096–1104 (2007).
Osman, F. & Whitby, M. C. Exploring the roles of Mus81-Eme1/Mms4 at perturbed replication forks. DNA Repair 6, 1004–1017 (2007).
Ho, S. S. et al. The DNA structure-specific endonuclease MUS81 mediates DNA sensor STING-dependent host rejection of prostate cancer cells. Immunity 44, 1177–1189 (2016).
Xiao, Y. et al. Comprehensive analysis of DNA damage repair deficiency in 10,284 pan-cancer study. Ann. Transl. Med. 9, 1661 (2021).
Härtlova, A. et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity 42, 332–343 (2015).
Lopez-Pelaez, M. et al. Targeting DNA damage response components induces enhanced STING-dependent type-I IFN response in ATM deficient cancer cells and drives dendritic cell activation. Oncoimmunology 11, 2117321 (2022).
Lu, C. et al. DNA sensing in mismatch repair-deficient tumor cells is essential for anti-tumor immunity. Cancer Cell 39, 96–108.e6 (2021).
Guan, J. et al. MLH1 deficiency-triggered DNA hyperexcision by exonuclease 1 activates the cGAS-STING pathway. Cancer Cell 39, 109–121.e105 (2021). This study, together with Lu et al. (2021), not only reveals the essential role of the tumour-intrinsic cGAS–STING pathway in establishing a ‘hot’ TME in MMR-deficient cancers and enhancing ICB efficiency, but also provides the molecular mechanism for how deficiency of MLH1 generates cytosolic DNA.
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413 (2017).
Bryan, T. M., Englezou, A., Dalla-Pozza, L., Dunham, M. A. & Reddel, R. R. Evidence for an alternative mechanism for maintaining telomere length in human tumors and tumor-derived cell lines. Nat. Med. 3, 1271–1274 (1997).
Kim, N. W. et al. Specific association of human telomerase activity with immortal cells and cancer. Science 266, 2011–2015 (1994).
Henson, J. et al. A robust assay for alternative lengthening of telomeres (ALT) in tumors demonstrates the significance of ALT in sarcomas and astrocytomas. Clin. Cancer Res. 11, 217–225 (2005).
Chen, Y.-A. et al. Extrachromosomal telomere repeat DNA is linked to ALT development via cGAS–STING DNA sensing pathway. Nat. Struct. Mol. Biol. 24, 1124–1131 (2017).
Mender, I. et al. Telomere stress potentiates STING-dependent anti-tumor immunity. Cancer Cell 38, 400–411.e406 (2020).
Turner, K. M. et al. Extrachromosomal oncogene amplification drives tumour evolution and genetic heterogeneity. Nature 543, 122–125 (2017).
Hung, K. L. et al. Coordinated inheritance of extrachromosomal DNAs in cancer cells. Nature 635, 201–209 (2024).
Wang, Y. et al. eccDNAs are apoptotic products with high innate immunostimulatory activity. Nature 599, 308–314 (2021).
Petermann, E., Lan, L. & Zou, L. Sources, resolution and physiological relevance of R-loops and RNA–DNA hybrids. Nat. Rev. Mol. Cell Biol. 23, 521–540 (2022).
Crossley, M. P. et al. R-loop-derived cytoplasmic RNA–DNA hybrids activate an immune response. Nature 613, 187–194 (2023).
Maxwell, M. B. et al. ARID1A suppresses R-loop-mediated STING-type I interferon pathway activation of anti-tumor immunity. Cell 187, 3390–3408.e19 (2024).
West, A. P. & Shadel, G. S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 17, 363–375 (2017).
Gulen, M. F. et al. cGAS–STING drives ageing-related inflammation and neurodegeneration. Nature 620, 374–380 (2023).
Rongvaux, A. et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 159, 1563–1577 (2014).
White, M. J. et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 159, 1549–1562 (2014). This study, together with Rongvaux et al. (2014), shows that apoptosis silences immune responses by degrading mitochondrial DNA, which otherwise could trigger the cGAS–STING–IFN signal.
Victorelli, S. et al. Apoptotic stress causes mtDNA release during senescence and drives the SASP. Nature 622, 627–636 (2023).
West, A. P. et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature 520, 553–557 (2015).
Saha, S. et al. Serine depletion promotes antitumor immunity by activating mitochondrial DNA-mediated cGAS–STING signaling. Cancer Res. 84, 2645–2659 (2024).
Han, C. et al. Tumor cells suppress radiation-induced immunity by hijacking caspase 9 signaling. Nat. Immunol. 21, 546–554 (2020).
Lei, Y. et al. Cooperative sensing of mitochondrial DNA by ZBP1 and cGAS promotes cardiotoxicity. Cell 186, 3013–3032.e3022 (2023).
Yamazaki, T. et al. Mitochondrial DNA drives abscopal responses to radiation that are inhibited by autophagy. Nat. Immunol. 21, 1160–1171 (2020).
Wang, C. et al. Manganese increases the sensitivity of the cGAS–STING pathway for double-stranded DNA and is required for the host defense against DNA viruses. Immunity 48, 675–687.e677 (2018).
Huang, A. & Zhou, W. Mn-based cGAS–STING activation for tumor therapy. Chin. J. Cancer Res. 35, 19–43 (2023).
Liu, H. P. et al. Nuclear cGAS suppresses DNA repair and promotes tumorigenesis. Nature 563, 131–136 (2018). This study demonstrates that nuclear cGAS promotes tumorigenesis by inhibiting homologous recombination repair in a STING-independent manner.
Jiang, H. et al. Chromatin-bound cGAS is an inhibitor of DNA repair and hence accelerates genome destabilization and cell death. EMBO J. 38, e102718 (2019).
Chen, H. et al. cGAS suppresses genomic instability as a decelerator of replication forks. Sci. Adv. 6, eabb8941 (2020).
Lv, G. et al. mTORC2-driven chromatin cGAS mediates chemoresistance through epigenetic reprogramming in colorectal cancer. Nat. Cell Biol. 26, 1585–1596 (2024).
Dunphy, G. et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol. Cell 71, 745–760.e745 (2018).
Zhang, L. T. et al. STING is a cell-intrinsic metabolic checkpoint restricting aerobic glycolysis by targeting HK2. Nat. Cell Biol. 25, 1208–1222 (2023).
Ranoa, D. R. E. et al. STING promotes homeostasis via regulation of cell proliferation and chromosomal stability. Cancer Res. 79, 1465–1479 (2019).
Kuilman, T., Michaloglou, C., Mooi, W. J. & Peeper, D. S. The essence of senescence. Genes Dev. 24, 2463–2479 (2010).
Xue, W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007).
d’Adda di Fagagna, F. Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522 (2008).
Gluck, S. et al. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 19, 1061–1070 (2017).
Yang, H., Wang, H., Ren, J., Chen, Q. & Chen, Z. J. cGAS is essential for cellular senescence. Proc. Natl Acad. Sci. USA 114, E4612–E4620 (2017).
Dou, Z. et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 550, 402–406 (2017). This study shows that senescence-mediated prevention of tumorigenesis is dependent on STING and further that cGAS or STING expression in human cancers correlates with tumour-promoting chronic inflammation, but not IFN-I signal.
Kang, T. W. et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature 479, 547–551 (2011).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Nassour, J. et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature 565, 659–663 (2019). This study demonstrates that the cGAS–STING pathway contributes to telomere-driven autophagy, which prevents tumorigenesis by inducing cell death of cultured fibroblasts.
Cho, M.-G. et al. MRE11 liberates cGAS from nucleosome sequestration during tumorigenesis. Nature 625, 585–592 (2024).
Woo, S. R. et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity 41, 830–842 (2014).
Mowat, C., Mosley, S. R., Namdar, A., Schiller, D. & Baker, K. Anti-tumor immunity in mismatch repair-deficient colorectal cancers requires type I IFN-driven CCL5 and CXCL10. J. Exp. Med. 218, e20210108 (2021).
Vornholz, L. et al. Synthetic enforcement of STING signaling in cancer cells appropriates the immune microenvironment for checkpoint inhibitor therapy. Sci. Adv. 9, eadd8564 (2023).
Acha-Sagredo, A. et al. A constitutive interferon-high immunophenotype defines response to immunotherapy in colorectal cancer. Cancer Cell 43, 292–307.e297 (2025).
Li, L., Li, M., Jiang, Z. & Wang, X. ARID1A mutations are associated with increased immune activity in gastrointestinal cancer. Cells 8, 678 (2019).
Okamura, R. et al. ARID1A alterations function as a biomarker for longer progression-free survival after anti-PD-1/PD-L1 immunotherapy. J. Immunother. Cancer 8, e000438 (2020).
Goswami, S. et al. ARID1A mutation plus CXCL13 expression act as combinatorial biomarkers to predict responses to immune checkpoint therapy in mUCC. Sci. Transl. Med. 12, eabc4220 (2020).
Phan, T. G. & Croucher, P. I. The dormant cancer cell life cycle. Nat. Rev. Cancer 20, 398–411 (2020).
Hu, J. et al. STING inhibits the reactivation of dormant metastasis in lung adenocarcinoma. Nature 616, 806–813 (2023). This study shows that tumour-cell-intrinsic STING restricts reactivation of dormant disseminated tumour cells in the lungs.
Chabanon, R. M. et al. Targeting the DNA damage response in immuno-oncology: developments and opportunities. Nat. Rev. Cancer 21, 701–717 (2021).
Yum, S., Li, M. & Chen, Z. J. Old dogs, new trick: classic cancer therapies activate cGAS. Cell Res. 30, 639–648 (2020).
Deng, L. et al. STING-dependent cytosolic DNA sensing promotes radiation-induced type I interferon-dependent antitumor immunity in immunogenic tumors. Immunity 41, 843–852 (2014). This study shows that the antitumour effects of radiation are dependent on STING-mediated DNA sensing, which elicits potent antitumour immunity.
Chen, D. et al. Tumor treating fields dually activate STING and AIM2 inflammasomes to induce adjuvant immunity in glioblastoma. J. Clin. Invest. 132, e149258 (2022).
Zierhut, C. et al. The cytoplasmic DNA sensor cGAS promotes mitotic cell death. Cell 178, 302–315.e323 (2019).
Ding, L. et al. PARP inhibition elicits STING-dependent antitumor immunity in Brca1-deficient ovarian cancer. Cell Rep. 25, 2972–2980.e2975 (2018).
Pantelidou, C. et al. PARP inhibitor efficacy depends on CD8(+) T-cell recruitment via intratumoral STING pathway activation in BRCA-deficient models of triple-negative breast cancer. Cancer Discov. 9, 722–737 (2019).
Chabanon, R. M. et al. PARP inhibition enhances tumor cell-intrinsic immunity in ERCC1-deficient non-small cell lung cancer. J. Clin. Invest. 129, 1211–1228 (2019).
Shen, J. et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res. 79, 311–319 (2019).
Ma, H., Kang, Z., Foo, T. K., Shen, Z. & Xia, B. Disrupted BRCA1–PALB2 interaction induces tumor immunosuppression and T-lymphocyte infiltration in HCC through cGAS–STING pathway. Hepatology 77, 33–47 (2023).
Kitai, Y. et al. DNA-containing exosomes derived from cancer cells treated with topotecan activate a STING-dependent pathway and reinforce antitumor immunity. J. Immunol. 198, 1649–1659 (2017).
Wang, Z. et al. cGAS/STING axis mediates a topoisomerase II inhibitor-induced tumor immunogenicity. J. Clin. Invest. 129, 4850–4862 (2019).
Wang, L. L. et al. Inhibition of the ATM/Chk2 axis promotes cGAS/STING signaling in ARID1A-deficient tumors. J. Clin. Invest. 130, 5951–5966 (2020).
Sen, T. et al. Targeting DNA damage response promotes antitumor immunity through STING-mediated T-cell activation in small cell lung cancer. Cancer Discov. 9, 646–661 (2019).
Kitajima, S. et al. MPS1 inhibition primes immunogenicity of KRAS-LKB1 mutant lung cancer. Cancer Cell 40, 1128–1144.e1128 (2022).
Parkes, E. E. et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J. Natl Cancer Inst. 109, djw199 (2017).
Lohard, S. et al. STING-dependent paracriny shapes apoptotic priming of breast tumors in response to anti-mitotic treatment. Nat. Commun. 11, 259 (2020).
Xu, M. M. et al. Dendritic cells but not macrophages sense tumor mitochondrial DNA for cross-priming through signal regulatory protein α signaling. Immunity 47, 363–373.e365 (2017).
Jneid, B. et al. Selective STING stimulation in dendritic cells primes antitumor T cell responses. Sci. Immunol. 8, eabn6612 (2023).
Jeong, S.-H., Yang, M. J., Choi, S., Kim, J. & Koh, G. Y. Refractoriness of STING therapy is relieved by AKT inhibitor through effective vascular disruption in tumour. Nat. Commun. 12, 4405 (2021).
Lam, K. C. et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 184, 5338–5356.e5321 (2021).
Marcus, A. et al. Tumor-derived cGAMP triggers a STING-mediated interferon response in non-tumor cells to activate the NK cell response. Immunity 49, 754–763.e754 (2018).
Nicolai, C. J. et al. NK cells mediate clearance of CD8+ T cell-resistant tumors in response to STING agonists. Sci. Immunol. 5, eaaz2738 (2020).
Lu, L. et al. STING signaling promotes NK cell antitumor immunity and maintains a reservoir of TCF-1+ NK cells. Cell Rep. 42, 113108 (2023).
Shacter, E. & Weitzman, S. A. Chronic inflammation and cancer. Oncology 16, 217–226 (2002).
Ahn, J. et al. Inflammation-driven carcinogenesis is mediated through STING. Nat. Commun. 5, 5166 (2014). This study reveals that carcinogen-induced skin tumorigenesis is dependent on STING, because its knockout in mice dramatically reduces number of tumours.
Chang, L., Guo, R., Huang, Q. & Yen, Y. Chromosomal instability triggered by Rrm2b loss leads to IL-6 secretion and plasmacytic neoplasms. Cell Rep. 3, 1389–1397 (2013).
Wörmann, S. M. et al. APOBEC3A drives deaminase domain-independent chromosomal instability to promote pancreatic cancer metastasis. Nat. Cancer 2, 1338–1356 (2021).
Zhang, H. et al. TMEM173 drives lethal coagulation in sepsis. Cell Host Microbe 27, 556–570.e556 (2020).
Smith, J. A. STING, the endoplasmic reticulum, and mitochondria: is three a crowd or a conversation? Front. Immunol. 11, 611347 (2021).
Chen, Q. et al. Carcinoma–astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature 533, 493–498 (2016).
Birkbak, N. J. et al. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 71, 3447–3452 (2011).
Wang, H. et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc. Natl Acad. Sci. USA 114, 1637–1642 (2017).
Corrales, L. et al. Direct activation of STING in the tumor microenvironment leads to potent and systemic tumor regression and immunity. Cell Rep. 11, 1018–1030 (2015).
Kong, X. et al. STING as an emerging therapeutic target for drug discovery: perspectives from the global patent landscape. J. Adv. Res. 44, 119–133 (2023).
Yum, S., Li, M., Frankel, A. E. & Chen, Z. J. Roles of the cGAS–STING pathway in cancer immunosurveillance and immunotherapy. Annu. Rev. Cancer Biol. 3, 323–344 (2019).
Lara, P. N. Jr et al. Randomized phase III placebo-controlled trial of carboplatin and paclitaxel with or without the vascular disrupting agent vadimezan (ASA404) in advanced non-small-cell lung cancer. J. Clin. Oncol. 29, 2965–2971 (2011).
Conlon, J. et al. Mouse, but not human STING, binds and signals in response to the vascular disrupting agent 5,6-dimethylxanthenone-4-acetic acid. J. Immunol. 190, 5216–5225 (2013).
Sivick, K. E. et al. Magnitude of therapeutic STING activation determines CD8(+) T cell-mediated anti-tumor immunity. Cell Rep. 29, 785–789 (2019).
Meric-Bernstam, F. et al. Phase I dose-escalation trial of MIW815 (ADU-S100), an intratumoral STING agonist, in patients with advanced/metastatic solid tumors or lymphomas. Clin. Cancer Res. 28, 677–688 (2022).
Zandberg, D. et al. 71PA phase II study of ADU-S100 in combination with pembrolizumab in adult patients with PD-L1+ recurrent or metastatic HNSCC: preliminary safety, efficacy and PK/PD results. Ann. Oncol. 31, S1446–S1447 (2020).
Harrington, K. et al. Preliminary results of the first-in-human (FIH) study of MK-1454, an agonist of stimulator of interferon genes (STING), as monotherapy or in combination with pembrolizumab (pembro) in patients with advanced solid tumors or lymphomas. Ann. Oncol. 29, viii712 (2018).
Wu, J., Dobbs, N., Yang, K. & Yan, N. Interferon-independent activities of mammalian STING mediate antiviral response and tumor immune evasion. Immunity 53, 115–126.e115 (2020).
Gulen, M. F. et al. Signalling strength determines proapoptotic functions of STING. Nat. Commun. 8, 427 (2017).
Cristescu, R. et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593 (2018).
Dosta, P., Cryer, A. M., Prado, M. & Artzi, N. Bioengineering strategies to optimize STING agonist therapy. Nat. Rev. Bioeng. 3, 660–680 (2025).
Zhang, Z. & Zhang, C. Regulation of cGAS–STING signalling and its diversity of cellular outcomes. Nat. Rev. Immunol. 25, 425–444 (2025).
Orzalli, M. H. et al. cGAS-mediated stabilization of IFI16 promotes innate signaling during herpes simplex virus infection. Proc. Natl Acad. Sci. USA 112, E1773–E1781 (2015).
Dvorkin, S., Cambier, S., Volkman, H. E. & Stetson, D. B. New frontiers in the cGAS–STING intracellular DNA-sensing pathway. Immunity 57, 718–730 (2024).
Guey, B. et al. BAF restricts cGAS on nuclear DNA to prevent innate immune activation. Science 369, 823–828 (2020).
Volkman, H. E., Cambier, S., Gray, E. E. & Stetson, D. B. Tight nuclear tethering of cGAS is essential for preventing autoreactivity. eLife 8, e47491 (2019).
Li, T. et al. Phosphorylation and chromatin tethering prevent cGAS activation during mitosis. Science 371, eabc5386 (2021).
Xu, P. et al. The CRL5–SPSB3 ubiquitin ligase targets nuclear cGAS for degradation. Nature 627, 873–879 (2024).
Barnett, K. C. et al. Phosphoinositide interactions position cGAS at the plasma membrane to ensure efficient distinction between self- and viral DNA. Cell 176, 1432–1446.e11 (2019).
Wang, H. et al. MYO1F positions cGAS on the plasma membrane to ensure full and functional signaling. Mol. Cell 85, 150–165.e157 (2025).
Willingham, S. B. et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl Acad. Sci. USA 109, 6662–6667 (2012).
Liu, X. et al. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat. Med. 21, 1209–1215 (2015).
de Mingo Pulido, Á et al. The inhibitory receptor TIM-3 limits activation of the cGAS–STING pathway in intra-tumoral dendritic cells by suppressing extracellular DNA uptake. Immunity 54, 1154–1167.e1157 (2021).
Dixon, K. O. et al. TIM-3 restrains anti-tumour immunity by regulating inflammasome activation. Nature 595, 101–106 (2021).
Xia, T., Konno, H., Ahn, J. & Barber, G. N. Deregulation of STING signaling in colorectal carcinoma constrains DNA damage responses and correlates with tumorigenesis. Cell Rep. 14, 282–297 (2016).
Konno, H. et al. Suppression of STING signaling through epigenetic silencing and missense mutation impedes DNA damage mediated cytokine production. Oncogene 37, 2037–2051 (2018).
Skoulidis, F. et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 8, 822–835 (2018).
Xia, T., Konno, H. & Barber, G. N. Recurrent loss of STING signaling in melanoma correlates with susceptibility to viral oncolysis. Cancer Res. 76, 6747–6759 (2016).
Falahat, R. et al. Epigenetic reprogramming of tumor cell–intrinsic STING function sculpts antigenicity and T cell recognition of melanoma. Proc. Natl Acad. Sci. USA 118, e2013598118 (2021).
Wu, M.-Z. et al. miR-25/93 mediates hypoxia-induced immunosuppression by repressing cGAS. Nat. Cell Biol. 19, 1286–1296 (2017).
Huang, B., Song, B. -l & Xu, C. Cholesterol metabolism in cancer: mechanisms and therapeutic opportunities. Nat. Metab. 2, 132–141 (2020).
Zhang, B. -c et al. Cholesterol-binding motifs in STING that control endoplasmic reticulum retention mediate anti-tumoral activity of cholesterol-lowering compounds. Nat. Commun. 15, 2760 (2024).
York, A. G. et al. Limiting cholesterol biosynthetic flux spontaneously engages type I IFN signaling. Cell 163, 1716–1729 (2015).
Ippolito, L., Morandi, A., Giannoni, E. & Chiarugi, P. Lactate: a metabolic driver in the tumour landscape. Trends Biochem. Sci. 44, 153–166 (2019).
Chen, H. et al. NBS1 lactylation is required for efficient DNA repair and chemotherapy resistance. Nature 631, 663–669 (2024).
Li, H. et al. AARS1 and AARS2 sense l-lactate to regulate cGAS as global lysine lactyltransferases. Nature 634, 1229–1237 (2024).
Gui, X. et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature 567, 262–266 (2019).
Zhao, M. et al. CGAS is a micronucleophagy receptor for the clearance of micronuclei. Autophagy 17, 3976–3991 (2021).
Konno, H., Konno, K. & Barber, G. N. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell 155, 688–698 (2013).
Prabakaran, T. et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM 1-dependent autophagy pathway activated by TBK1. EMBO J. 37, e97858 (2018).
Liu, D. et al. STING directly activates autophagy to tune the innate immune response. Cell Death Differ. 26, 1735–1749 (2019).
Mohr, L. et al. ER-directed TREX1 limits cGAS activation at micronuclei. Mol. Cell 81, 724–738.e729 (2021).
Vanpouille-Box, C. et al. DNA exonuclease Trex1 regulates radiotherapy-induced tumour immunogenicity. Nat. Commun. 8, 15618 (2017).
Tani, T. et al. TREX1 inactivation unleashes cancer cell STING–interferon signaling and promotes antitumor immunity. Cancer Discov. 14, 752–765 (2024).
Ghosh, M., Saha, S., Li, J., Montrose, D. C. & Martinez, L. A. p53 engages the cGAS/STING cytosolic DNA sensing pathway for tumor suppression. Mol. Cell 83, 266–280.e266 (2023).
Salojin, C. et al. 765 The first-in-class small molecule TREX1 inhibitor CPI-381 demonstrates type I IFN induction and sensitization of tumors to immune checkpoint blockade. J. Immunother. Cancer 9, A800–A800 (2021).
Carozza, J. A. et al. Extracellular cGAMP is a cancer-cell-produced immunotransmitter involved in radiation-induced anticancer immunity. Nat. Cancer 1, 184–196 (2020).
Li, L. et al. Hydrolysis of 2′ 3′-cGAMP by ENPP1 and design of nonhydrolyzable analogs. Nat. Chem. Biol. 10, 1043–1048 (2014).
Mardjuki, R. et al. Identification of the extracellular membrane protein ENPP3 as a major cGAMP hydrolase and innate immune checkpoint. Cell Rep. 43, 114209 (2024).
Hou, Y. F. et al. SMPDL3A is a cGAMP-degrading enzyme induced by LXR-mediated lipid metabolism to restrict cGAS–STING DNA sensing. Immunity 56, 2492–2507.e10 (2023).
Carozza, J. A. et al. Structure-aided development of small-molecule inhibitors of ENPP1, the extracellular phosphodiesterase of the immunotransmitter cGAMP. Cell Chem. Biol. 27, 1347–1358.e1345 (2020).
Ghosh, M. et al. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell 39, 494–508.e5 (2021).
Hou, Y. et al. Non-canonical NF-κB antagonizes STING sensor-mediated DNA sensing in radiotherapy. Immunity 49, 490–503.e494 (2018).
Nakajima, S. et al. Radiation-induced remodeling of the tumor microenvironment through tumor cell-intrinsic expression of cGAS–STING in esophageal squamous cell carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 115, 957–971 (2023).
Li, S. et al. STING-induced regulatory B cells compromise NK function in cancer immunity. Nature 610, 373–380 (2022).
Liang, H. et al. Host STING-dependent MDSC mobilization drives extrinsic radiation resistance. Nat. Commun. 8, 1736 (2017).
Song, H. et al. Targeting tumor monocyte-intrinsic PD-L1 by rewiring STING signaling and enhancing STING agonist therapy. Cancer Cell 43, 503–518.e510 (2025).
Lemos, H. et al. STING promotes the growth of tumors characterized by low antigenicity via IDO activation. Cancer Res. 76, 2076–2081 (2016).
Pang, E. S. et al. Discordance in STING-induced activation and cell death between mouse and human dendritic cell populations. Front. Immunol. 13, 794776 (2022).
Gaidt, M. M. et al. The DNA inflammasome in human myeloid cells is initiated by a STING-cell death program upstream of NLRP3. Cell 171, 1110–1124.e1118 (2017).
Li, W. et al. cGAS–STING-mediated DNA sensing maintains CD8(+) T cell stemness and promotes antitumor T cell therapy. Sci. Transl. Med. 12, eaay9013 (2020).
Wu, J. et al. STING-mediated disruption of calcium homeostasis chronically activates ER stress and primes T cell death. J. Exp. Med. 216, 867–883 (2019).
Cerboni, S. et al. Intrinsic antiproliferative activity of the innate sensor STING in T lymphocytes. J. Exp. Med. 214, 1769–1785 (2017).
Larkin, B. et al. Cutting edge: activation of STING in T cells induces type I IFN responses and cell death. J. Immunol. 199, 397–402 (2017).
Kuhl, N. et al. STING agonism turns human T cells into interferon-producing cells but impedes their functionality. EMBO Rep. 24, e55536 (2023).
Boukhaled, G. M. et al. Pre-encoded responsiveness to type I interferon in the peripheral immune system defines outcome of PD1 blockade therapy. Nat. Immunol. 23, 1273–1283 (2022).
Gandhi, L. et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N. Engl. J. Med. 378, 2078–2092 (2018).
Shitara, K. et al. Efficacy and safety of pembrolizumab or pembrolizumab plus chemotherapy vs chemotherapy alone for patients with first-line, advanced gastric cancer: the KEYNOTE-062 phase 3 randomized clinical trial. JAMA Oncol. 6, 1571–1580 (2020).
Lee, N. Y. et al. Avelumab plus standard-of-care chemoradiotherapy versus chemoradiotherapy alone in patients with locally advanced squamous cell carcinoma of the head and neck: a randomised, double-blind, placebo-controlled, multicentre, phase 3 trial. Lancet Oncol. 22, 450–462 (2021).
Machiels, J.-P. et al. Pembrolizumab plus concurrent chemoradiotherapy versus placebo plus concurrent chemoradiotherapy in patients with locally advanced squamous cell carcinoma of the head and neck (KEYNOTE-412): a randomised, double-blind, phase 3 trial. Lancet Oncol. 25, 572–587 (2024).
Grassberger, C., Ellsworth, S. G., Wilks, M. Q., Keane, F. K. & Loeffler, J. S. Assessing the interactions between radiotherapy and antitumour immunity. Nat. Rev. Clin. Oncol. 16, 729–745 (2019).
Ogitani, Y. et al. DS-8201a, a novel HER2-targeting ADC with a novel DNA topoisomerase I inhibitor, demonstrates a promising antitumor efficacy with differentiation from T-DM1. Clin. Cancer Res. 22, 5097–5108 (2016).
Powles, T. et al. Enfortumab vedotin and pembrolizumab in untreated advanced urothelial cancer. N. Engl. J. Med. 390, 875–888 (2024).
Powles, T. et al. Pembrolizumab alone or combined with chemotherapy versus chemotherapy as first-line therapy for advanced urothelial carcinoma (KEYNOTE-361): a randomised, open-label, phase 3 trial. Lancet Oncol. 22, 931–945 (2021).
Wu, Y.-t. et al. Tumor-targeted delivery of a STING agonist improves cancer immunotherapy. Proc. Natl Acad. Sci. USA 119, e2214278119 (2022).
Soomer-James, J. T., Lancaster, K., Damelin, M. & Malli, N. XMT-2056, a HER2-directed STING agonist antibody–drug conjugate, exhibits ADCC function that synergizes with STING pathway activation and contributes to anti-tumor responses. Cancer Res. 83, 4423 (2023).
Cetinbas, N. M. et al. Tumor cell-directed STING agonist antibody–drug conjugates induce type III interferons and anti-tumor innate immune responses. Nat. Commun. 15, 5842 (2024).
US National Library of Medicine. ClinicalTrials.gov http://www.clinicaltrials.gov/show/NCT05514717 (2025).
Lin, W. et al. STING trafficking activates MAPK–CREB signaling to trigger regulatory T cell differentiation. Proc. Natl Acad. Sci. USA 121, e2320709121 (2024).
Liu, B. et al. Human STING is a proton channel. Science 381, 508–514 (2023). This study reveals that STING can transport protons across membranes, resulting in LC3B lipidation and inflammasome activation.
Xun, J. et al. A conserved ion channel function of STING mediates noncanonical autophagy and cell death. EMBO Rep. 25, 544–569 (2024).
Lv, B. et al. A TBK1-independent primordial function of STING in lysosomal biogenesis. Mol. Cell 84, 3979–3996.e3979 (2024).
Xu, Y. et al. The cGAS–STING pathway activates transcription factor TFEB to stimulate lysosome biogenesis and pathogen clearance. Immunity 58, 309–325.e6 (2024).
Huang, T., Sun, C., Du, F. & Chen, Z. J. STING-induced noncanonical autophagy regulates endolysosomal homeostasis. Proc. Natl Acad. Sci. USA 122, e2415422122 (2025).
Poddar, S. et al. ArfGAP2 promotes STING proton channel activity, cytokine transit, and autoinflammation. Cell 188, 1605–1622.e26 (2025).
Wang, Y. et al. Universal STING mimic boosts antitumour immunity via preferential activation of tumour control signalling pathways. Nat. Nanotechnol. 19, 856–886 (2024).
Pao, W. et al. Tissue-specific immunoregulation: a call for better understanding of the “immunostat” in the context of cancer. Cancer Discov. 8, 395–402 (2018).
Siddiqui, I. et al. Intratumoral Tcf1+ PD-1+ CD8+ T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity 50, 195–211.e110 (2019).
Kurtulus, S. et al. Checkpoint blockade immunotherapy induces dynamic changes in PD-1− CD8+ tumor-infiltrating T cells. Immunity 50, 181–194.e186 (2019).
Escobar, G. et al. Tumor immunogenicity dictates reliance on TCF1 in CD8+ T cells for response to immunotherapy. Cancer Cell 41, 1662–1679.e1667 (2023).
Li, Z. et al. Novel photo-STING agonists delivered by erythrocyte efferocytosis-mimicking pattern to repolarize tumor-associated macrophages for boosting anticancer immunotherapy. Adv. Mater. 36, 2410937 (2024).
Wei, X. et al. LL-37 transports immunoreactive cGAMP to activate STING signaling and enhance interferon-mediated host antiviral immunity. Cell Rep. 39, 110880 (2022).
Zhang, Q. et al. Molecular basis of SLC19A1-mediated folate and cyclic dinucleotide transport. Nat. Commun. 16, 3146 (2025).
Ding, Z. et al. Single atom engineering for radiotherapy-activated immune agonist prodrugs. Nat. Commun. 16, 6021 (2025).
Yin, X. et al. Orchestrating intratumoral DC-T cell immunity for enhanced tumor control via radiotherapy-activated TLR7/8 prodrugs in mice. Nat. Commun. 16, 6020 (2025).
Jiang, J. et al. Sono-driven STING activation using semiconducting polymeric nanoagonists for precision sono-immunotherapy of head and neck squamous cell carcinoma. Adv. Mater. 35, 2300854 (2023).
Baxter, A. G. & Hodgkin, P. D. Activation rules: the two-signal theories of immune activation. Nat. Rev. Immunol. 2, 439–446 (2002).
Chen, L. Co-inhibitory molecules of the B7–CD28 family in the control of T-cell immunity. Nat. Rev. Immunol. 4, 336–347 (2004).
Heemskerk, B., Kvistborg, P. & Schumacher, T. N. M. The cancer antigenome. EMBO J. 32, 194–203 (2013).
Maleki Vareki, S. High and low mutational burden tumors versus immunologically hot and cold tumors and response to immune checkpoint inhibitors. J. Immunother. Cancer 6, 157 (2018).
Tang, H. D. et al. Facilitating T cell infiltration in tumor microenvironment overcomes resistance to PD-L1 blockade. Cancer Cell 29, 285–296 (2016).
Tumeh, P. C. et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515, 568–571 (2014).
Greten, F. R. & Grivennikov, S. I. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity 51, 27–41 (2019).
Zak, J. et al. JAK inhibition enhances checkpoint blockade immunotherapy in patients with Hodgkin lymphoma. Science 384, eade8520 (2024).
Mathew, D. et al. Combined JAK inhibition and PD-1 immunotherapy for non-small cell lung cancer patients. Science 384, eadf1329 (2024). This study, together with Zak et al. (2024), reveals that JAK1 and JAK2 are a target of chronic inflammation and transient blockade of JAK1/2 signal enhances T cell function and synergizes with anti-PD1 in both mouse model and patients with cancers.
Yu, Y. et al. Post-translational modifications of cGAS–STING: a critical switch for immune regulation. Cells 11, 3043 (2022).
Kang, J. et al. Post-translational modifications of STING: a potential therapeutic target. Front. Immunol. 13, 888147 (2022).
Seo, G. J. et al. Akt kinase-mediated checkpoint of cGAS DNA sensing pathway. Cell Rep. 13, 440–449 (2015).
Zhong, L. et al. Phosphorylation of cGAS by CDK1 impairs self-DNA sensing in mitosis. Cell Discov. 6, 26 (2020).
Zhang, C. et al. Structural basis of STING binding with and phosphorylation by TBK1. Nature 567, 394–398 (2019).
Liu, S. et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347, aaa2630 (2015).
Ma, M. et al. TAK1 is an essential kinase for STING trafficking. Mol. Cell 83, 3885–3903.e3885 (2023).
Wang, C. et al. EGFR-mediated tyrosine phosphorylation of STING determines its trafficking route and cellular innate immunity functions. EMBO J. 39, e104106 (2020).
Seo, G. J. et al. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat. Commun. 9, 613 (2018).
Wang, Q. et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathog. 13, e1006264 (2017).
Chen, X. & Chen, Y. Ubiquitination of cGAS by TRAF6 regulates anti-DNA viral innate immune responses. Biochem. Biophys. Res. Commun. 514, 659–664 (2019).
Tsuchida, T. et al. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33, 765–776 (2010).
Zhang, J., Hu, M.-M., Wang, Y.-Y. & Shu, H.-B. TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287, 28646–28655 (2012).
Ni, G., Konno, H. & Barber, G. N. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci. Immunol. 2, eaah7119 (2017).
Zhang, Z.-D. et al. RNF115 plays dual roles in innate antiviral responses by catalyzing distinct ubiquitination of MAVS and MITA. Nat. Commun. 11, 5536 (2020).
Xing, J. et al. TRIM29 promotes DNA virus infections by inhibiting innate immune response. Nat. Commun. 8, 945 (2017).
Wang, Y. et al. TRIM30α is a negative-feedback regulator of the intracellular DNA and DNA virus-triggered response by targeting STING. PLoS Pathog. 11, e1005012 (2015).
Hu, M.-M. et al. Sumoylation promotes the stability of the DNA sensor cGAS and the adaptor STING to regulate the kinetics of response to DNA virus. Immunity 45, 555–569 (2016).
Cui, Y. et al. SENP7 potentiates cGAS activation by relieving SUMO-mediated inhibition of cytosolic DNA sensing. PLoS Pathog. 13, e1006156 (2017).
Li, C. et al. RNF111-facilitated neddylation potentiates cGAS-mediated antiviral innate immune response. PLoS Pathog. 17, e1009401 (2021).
Song, Z.-M. et al. KAT5 acetylates cGAS to promote innate immune response to DNA virus. Proc. Natl Acad. Sci. USA 117, 21568–21575 (2020).
Dai, J. et al. Acetylation blocks cGAS activity and inhibits self-DNA-induced autoimmunity. Cell 176, 1447–1460.e1414 (2019).
Ma, D. et al. Arginine methyltransferase PRMT5 negatively regulates cGAS-mediated antiviral immune response. Sci. Adv. 7, eabc1834 (2021).
Xia, P. et al. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 17, 369–378 (2016).
Shi, C. et al. ZDHHC18 negatively regulates cGAS-mediated innate immunity through palmitoylation. EMBO J. 41, e109272 (2022).
Mukai, K. et al. Activation of STING requires palmitoylation at the Golgi. Nat. Commun. 7, 11932 (2016).
Tao, L. et al. Reactive oxygen species oxidize STING and suppress interferon production. eLife 9, e57837 (2020).
Jia, M. et al. Redox homeostasis maintained by GPX4 facilitates STING activation. Nat. Immunol. 21, 727–735 (2020).
Wang, Y. et al. Inflammasome activation triggers caspase-1-mediated cleavage of cGAS to regulate responses to DNA virus infection. Immunity 46, 393–404 (2017).
Ning, X. et al. Apoptotic caspases suppress type I interferon production via the cleavage of cGAS, MAVS, and IRF3. Mol. Cell 74, 19–31.e17 (2019).
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (grant 82250710684 to Y.-X.F.), the Shenzhen Science and Technology Program (grant KQTD20240729102213019 to C.L.) and by the Major Program of Shenzhen Bay Laboratory (grants S241101007 and S201101004 to C.L.).
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Cancer thanks David Barbie, Samuel Bakhoum and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Abscopal tumours
-
A phenomenon where local treatment (such as radiation) of one tumour causes the regression of distant, untreated tumours.
- Autophagy
-
A conserved cellular catabolic process that degrades dysfunctional organelles and proteins within lysosomes, enabling nutrient recycling and maintenance of cellular homeostasis.
- Chromosomal instability
-
Persistent chromosome segregation errors during consecutive cell divisions, frequently leading to aneuploidy and contributing to tumour heterogeneity and drug resistance.
- DNA strand invasion
-
A process in DNA repair where a single-strand DNA invades a homologous double-strand DNA template, with defects in this process contributing to genomic instability and tumorigenesis.
- Immune checkpoint blockade
-
Therapeutic inhibition of checkpoint proteins (such as PD1, PDL1 and CTLA4) to enhance antitumour immune responses by reactivating T cells.
- Lactylation
-
A novel post-translational modification where a lactate molecule is covalently attached to a lysine residue on a protein, thereby altering the protein’s function and expression level.
- LC3 lipidation
-
The attachment of LC3 to phosphatidylethanolamine during autophagy, marking the formation of autophagosomes and serving as a key indicator of autophagic activity.
- M2-like macrophage
-
A macrophage that supports tissue repair, immune suppression and tumour progression by secreting anti-inflammatory cytokines such as IL-10 and TGFβ.
- Micronuclei
-
Membrane-bound structures containing lagging chromosomes that fail to integrate into daughter-cell nuclei during cell division, which indicates genomic instability.
- Necroptosis
-
A form of regulated cell death triggered by RIPK3 and its substrate MLKL, causing cell swelling, plasma membrane rupture, and the release of pro-inflammatory factors.
- Neoantigens
-
Tumour-specific antigens generated from somatic mutations, presented by MHC molecules, making them potential targets for immunotherapies such as cancer vaccines and T cell therapies.
- Proliferative crisis
-
A catastrophic state following replicative senescence, where cells with critically shortened telomeres continue to divide, resulting in genomic instability and massive cell death.
- Replication fork
-
The structure formed during DNA replication where the DNA double helix is unwound and new strands are synthesized, with stalling or collapse causing DNA damage, a hallmark of genomic instability.
- R-loop
-
A three-stranded structure formed when RNA displaces the non-template DNA strand, causing DNA damage and contributing to genomic instability.
- Senescence
-
A state of irreversible cell cycle arrest caused by DNA damage or telomere shortening, in which cells remain metabolically active but no longer divide, acting as a potential tumour-suppressive mechanism.
- SPNM nanoparticles
-
Smart polymeric nanoparticles designed for targeted drug delivery, that respond to specific stimuli (such as pH changes) to release drugs at tumour sites, thereby improving bioavailability and minimizing systemic side effects.
- Tumour treating fields
-
A non-invasive therapy that utilizes low-intensity, intermediate-frequency alternating electric fields to disrupt the normal polymerization and function of microtubules during mitosis, leading to mitotic arrest and programmed cell death in rapidly dividing cancer cells.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lu, C., Wang, W. & Fu, YX. Opportunities and challenges of targeting cGAS–STING in cancer. Nat Rev Cancer (2026). https://doi.org/10.1038/s41568-025-00894-9
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41568-025-00894-9