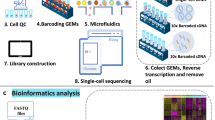
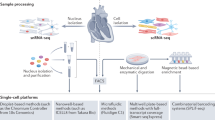
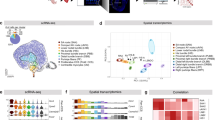
Abstract
The cardiac conduction system (CCS) has a vital role in initiating and coordinating nearly 3 billion heartbeats throughout a person’s lifetime. The CCS comprises two primary tissue types: the impulse-generating, slow-conducting nodes and the fast-conducting components of the ventricular conduction system. Dysfunction in this system can give rise to a spectrum of clinical symptoms, including palpitations, syncope, heart failure and even sudden cardiac death. Owing to the limited therapeutic options other than electronic pacemakers, substantial research efforts have been aimed at uncovering the root causes of conduction system disorders. A comprehensive investigative approach integrating genetics, transcriptomics and proteomics has been used to unravel the complex biology of these diseases. Advances in single-cell genomic and transcriptomic technologies, together with spatial transcriptomics, are offering new insights into the cellular microenvironments that govern conduction system function. In this Review, we examine the latest progress in understanding the biology of the CCS, situating new findings within both established and emerging scientific paradigms. Additionally, we discuss how these insights can be leveraged to improve clinical risk assessment, expand drug discovery efforts, accelerate technology aimed at promoting CCS regeneration and foster the development of innovative therapies, including biological pacemakers.
Key points
-
The cardiac conduction system (CCS) initiates each heartbeat and regulates the flow of electrical impulses throughout the heart.
-
Diseases of the CCS are a major cause of morbidity and mortality worldwide; research into therapeutic alternatives to electronic pacemakers is driving new discoveries in CCS biology.
-
CCS-restricted gene regulatory networks reinforce specialized conduction phenotypes and simultaneously repress working cardiomyocyte-specific programming.
-
Transcriptomic analyses at single-cell resolution, combined with spatial transcriptomics, are offering unprecedented insights into the local tissue microenvironments of the CCS and are driving advances in next-generation tools for drug discovery and therapies specifically targeting the conduction system.
-
Development of biological pacemakers that use transcription factors to reprogramme working cardiomyocytes into CCS cells holds substantial therapeutic potential.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Kusumoto, F. M. et al. 2018 ACC/AHA/HRS guideline on the evaluation and management of patients with bradycardia and cardiac conduction delay: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation 140, e382–e482 (2019).
Haimovich, J. S. et al. Frequency of electrocardiogram-defined cardiac conduction disorders in a multi-institutional primary care cohort. JACC Adv. 3, 101004 (2024).
Weng, L. C. et al. The impact of common and rare genetic variants on bradyarrhythmia development. Nat. Genet. 57, 53–64 (2025).
Milanesi, R., Baruscotti, M., Gnecchi-Ruscone, T. & DiFrancesco, D. Familial sinus bradycardia associated with a mutation in the cardiac pacemaker channel. N. Engl. J. Med. 354, 151–157 (2006).
Basson, C. T. et al. Mutations in human TBX5 [corrected] cause limb and cardiac malformation in Holt–Oram syndrome. Nat. Genet. 15, 30–35 (1997).
Crasto, S., My, I. & Di Pasquale, E. The broad spectrum of LMNA cardiac diseases: from molecular mechanisms to clinical phenotype. Front. Physiol. 11, 761 (2020).
Mond, H. G. & Proclemer, A. The 11th world survey of cardiac pacing and implantable cardioverter-defibrillators: calendar year 2009 — a World society of Arrhythmia’s project. Pacing Clin. Electrophysiol. 34, 1013–1027 (2011).
Simpson, J. et al. Long-term complications related to cardiac implantable electronic devices. J. Clin. Med. 14, 2058 (2025).
Pallante, B. A. et al. Contactin-2 expression in the cardiac Purkinje fiber network. Circ. Arrhythm. Electrophysiol. 3, 186–194 (2010).
Gillers, B. S. et al. Canonical wnt signaling regulates atrioventricular junction programming and electrophysiological properties. Circ. Res. 116, 398–406 (2015).
Nerbonne, J. M. & Kass, R. S. Molecular physiology of cardiac repolarization. Physiol. Rev. 85, 1205–1253 (2005).
Lakatta, E. G., Maltsev, V. A. & Vinogradova, T. M. A coupled system of intracellular Ca2+ clocks and surface membrane voltage clocks controls the timekeeping mechanism of the heart’s pacemaker. Circ. Res. 106, 659–673 (2010).
Linscheid, N. et al. Quantitative proteomics and single-nucleus transcriptomics of the sinus node elucidates the foundation of cardiac pacemaking. Nat. Commun. 10, 2889 (2019).
DiFrancesco, D. The contribution of the ‘pacemaker’ current (if) to generation of spontaneous activity in rabbit sino-atrial node myocytes. J. Physiol. 434, 23–40 (1991).
Kanemaru, K. et al. Spatially resolved multiomics of human cardiac niches. Nature 619, 801–810 (2023).
Platzer, J. et al. Congenital deafness and sinoatrial node dysfunction in mice lacking class D L-type Ca2+ channels. Cell 102, 89–97 (2000).
Mangoni, M. E. et al. Bradycardia and slowing of the atrioventricular conduction in mice lacking CaV3.1/alpha1G T-type calcium channels. Circ. Res. 98, 1422–1430 (2006).
Goodyer, W. R. et al. In vivo visualization and molecular targeting of the cardiac conduction system. J. Clin. Invest. 132, e156955 (2022).
Goodyer, W. R. et al. Transcriptomic profiling of the developing cardiac conduction system at single-cell resolution. Circ. Res. 125, 379–397 (2019).
Ivanov, S. V. et al. Cerebellar ataxia, seizures, premature death, and cardiac abnormalities in mice with targeted disruption of the Cacna2d2 gene. Am. J. Pathol. 165, 1007–1018 (2004).
Bogdanov, K. Y., Vinogradova, T. M. & Lakatta, E. G. Sinoatrial nodal cell ryanodine receptor and Na(+)-Ca(2+) exchanger: molecular partners in pacemaker regulation. Circ. Res. 88, 1254–1258 (2001).
Kreuzberg, M. M. et al. Connexin30.2 containing gap junction channels decelerate impulse propagation through the atrioventricular node. Proc. Natl Acad. Sci. USA 103, 5959–5964 (2006).
Aanhaanen, W. T. et al. Developmental origin, growth, and three-dimensional architecture of the atrioventricular conduction axis of the mouse heart. Circ. Res. 107, 728–736 (2010).
Saez, J. C., Berthoud, V. M., Branes, M. C., Martinez, A. D. & Beyer, E. C. Plasma membrane channels formed by connexins: their regulation and functions. Physiol. Rev. 83, 1359–1400 (2003).
Al-Khatib, S. M. et al. Risk stratification for arrhythmic events in patients with asymptomatic pre-excitation: a systematic review for the 2015 ACC/AHA/HRS guideline for the management of adult patients with supraventricular tachycardia: a report of the American College of Cardiology/American Heart Association Task Force on clinical practice guidelines and the Heart Rhythm Society. J. Am. Coll. Cardiol. 67, 1624–1638 (2016).
Hiroi, Y. et al. Tbx5 associates with Nkx2-5 and synergistically promotes cardiomyocyte differentiation. Nat. Genet. 28, 276–280 (2001).
Hoogaars, W. M. et al. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc. Res. 62, 489–499 (2004).
Linden, H., Williams, R., King, J., Blair, E. & Kini, U. Ulnar mammary syndrome and TBX3: expanding the phenotype. Am. J. Med. Genet. A 149A, 2809–2812 (2009).
Meneghini, V., Odent, S., Platonova, N., Egeo, A. & Merlo, G. R. Novel TBX3 mutation data in families with ulnar-mammary syndrome indicate a genotype–phenotype relationship: mutations that do not disrupt the T-domain are associated with less severe limb defects. Eur. J. Med. Genet. 49, 151–158 (2006).
Stennard, F. A. & Harvey, R. P. T-box transcription factors and their roles in regulatory hierarchies in the developing heart. Development 132, 4897–4910 (2005).
Liu, N. et al. Functional variants in TBX2 are associated with a syndromic cardiovascular and skeletal developmental disorder. Hum. Mol. Genet. 27, 2454–2465 (2018).
Bruneau, B. G. et al. A murine model of Holt–Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell 106, 709–721 (2001).
Garg, V. et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature 424, 443–447 (2003).
Luna-Zurita, L. et al. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell 164, 999–1014 (2016).
Christoffels, V. M. et al. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev. Dyn. 229, 763–770 (2004).
Boogerd, K. J. et al. Msx1 and Msx2 are functional interacting partners of T-box factors in the regulation of Connexin43. Cardiovasc. Res. 78, 485–493 (2008).
Hoogaars, W. M. et al. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 21, 1098–1112 (2007).
Habets, P. E. et al. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 16, 1234–1246 (2002).
Burnicka-Turek, O. et al. Coordinated Tbx3/Tbx5 transcriptional control of the adult ventricular conduction system. eLife 13, RP102027 (2025).
Schott, J. J. et al. Congenital heart disease caused by mutations in the transcription factor NKX2-5. Science 281, 108–111 (1998).
Oh, Y. et al. Transcriptional regulation of the postnatal cardiac conduction system heterogeneity. Nat. Commun. 15, 6550 (2024).
Wiesinger, A. et al. A single cell transcriptional roadmap of human pacemaker cell differentiation. eLife 11, e76781 (2022).
Bhattacharyya, S. et al. Using Gjd3-CreEGFP mice to examine atrioventricular node morphology and composition. Sci. Rep. 9, 2106 (2019).
Liang, D. et al. Cellular and molecular landscape of mammalian sinoatrial node revealed by single-cell RNA sequencing. Nat. Commun. 12, 287 (2021).
Farah, E. N. et al. Spatially organized cellular communities form the developing human heart. Nature 627, 854–864 (2024).
Arduini, A. et al. Transcriptional profile of the rat cardiovascular system at single-cell resolution. Cell Rep. 44, 115091 (2025).
van Eif, V. W. W. et al. Transcriptome analysis of mouse and human sinoatrial node cells reveals a conserved genetic program. Development 146, dev173161 (2019).
Kalyanasundaram, A. et al. Three-dimensional functional anatomy of the human sinoatrial node for epicardial and endocardial mapping and ablation. Heart Rhythm 20, 122–133 (2023).
Csepe, T. A. et al. Human sinoatrial node structure: 3D microanatomy of sinoatrial conduction pathways. Prog. Biophys. Mol. Biol. 120, 164–178 (2016).
Mommersteeg, M. T. et al. The sinus venosus progenitors separate and diversify from the first and second heart fields early in development. Cardiovasc. Res. 87, 92–101 (2010).
Mommersteeg, M. T. et al. Molecular pathway for the localized formation of the sinoatrial node. Circ. Res. 100, 354–362 (2007).
Wiese, C. et al. Formation of the sinus node head and differentiation of sinus node myocardium are independently regulated by Tbx18 and Tbx3. Circ. Res. 104, 388–397 (2009).
Mori, A. D. et al. Tbx5-dependent rheostatic control of cardiac gene expression and morphogenesis. Dev. Biol. 297, 566–586 (2006).
Blaschke, R. J. et al. Targeted mutation reveals essential functions of the homeodomain transcription factor Shox2 in sinoatrial and pacemaking development. Circulation 115, 1830–1838 (2007).
Ye, W. et al. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development 142, 2521–2532 (2015).
Espinoza-Lewis, R. A. et al. Ectopic expression of Nkx2.5 suppresses the formation of the sinoatrial node in mice. Dev. Biol. 356, 359–369 (2011).
Li, H. et al. Nkx2-5 defines a subpopulation of pacemaker cells and is essential for the physiological function of the sinoatrial node in mice. Development 146, dev178145 (2019).
Puskaric, S. et al. Shox2 mediates Tbx5 activity by regulating Bmp4 in the pacemaker region of the developing heart. Hum. Mol. Genet. 19, 4625–4633 (2010).
Basson, C. T. et al. The clinical and genetic spectrum of the Holt–Oram syndrome (heart-hand syndrome). N. Engl. J. Med. 330, 885–891 (1994).
Moskowitz, I. P. et al. The T-box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development 131, 4107–4116 (2004).
Mohan, R. A. et al. Embryonic Tbx3(+) cardiomyocytes form the mature cardiac conduction system by progressive fate restriction. Development 145, dev167361 (2018).
Vedantham, V., Galang, G., Evangelista, M., Deo, R. C. & Srivastava, D. RNA sequencing of mouse sinoatrial node reveals an upstream regulatory role for Islet-1 in cardiac pacemaker cells. Circ. Res. 116, 797–803 (2015).
Frank, D. U. et al. Lethal arrhythmias in Tbx3-deficient mice reveal extreme dosage sensitivity of cardiac conduction system function and homeostasis. Proc. Natl Acad. Sci. USA 109, E154–E163 (2012).
Mohan, R. A. et al. T-box transcription factor 3 governs a transcriptional program for the function of the mouse atrioventricular conduction system. Proc. Natl Acad. Sci. USA 117, 18617–18626 (2020).
Wu, M. et al. Baf250a orchestrates an epigenetic pathway to repress the Nkx2.5-directed contractile cardiomyocyte program in the sinoatrial node. Cell Res. 24, 1201–1213 (2014).
Gharibeh, L. et al. GATA6 is a regulator of sinus node development and heart rhythm. Proc. Natl Acad. Sci. USA 118, e2007322118 (2021).
Kapoor, N., Galang, G., Marban, E. & Cho, H. C. Transcriptional suppression of connexin43 by TBX18 undermines cell–cell electrical coupling in postnatal cardiomyocytes. J. Biol. Chem. 286, 14073–14079 (2011).
Kapoor, N., Liang, W., Marban, E. & Cho, H. C. Direct conversion of quiescent cardiomyocytes to pacemaker cells by expression of Tbx18. Nat. Biotechnol. 31, 54–62 (2013).
Wolfson, D. W. et al. Transient pacing in pigs with complete heart block via myocardial injection of mRNA coding for the T-box transcription factor 18. Nat. Biomed. Eng. 8, 1124–1141 (2024).
Espinoza-Lewis, R. A. et al. Shox2 is essential for the differentiation of cardiac pacemaker cells by repressing Nkx2-5. Dev. Biol. 327, 376–385 (2009).
Hoffmann, S. et al. Functional characterization of rare variants in the SHOX2 gene identified in sinus node dysfunction and atrial fibrillation. Front. Genet. 10, 648 (2019).
Ionta, V. et al. SHOX2 overexpression favors differentiation of embryonic stem cells into cardiac pacemaker cells, improving biological pacing ability. Stem Cell Rep. 4, 129–142 (2015).
Hoffmann, S. et al. Islet1 is a direct transcriptional target of the homeodomain transcription factor Shox2 and rescues the Shox2-mediated bradycardia. Basic Res. Cardiol. 108, 339 (2013).
Cai, C. L. et al. Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877–889 (2003).
Liang, X. et al. Transcription factor ISL1 is essential for pacemaker development and function. J. Clin. Invest. 125, 3256–3268 (2015).
Viragh, S. & Challice, C. E. The development of the conduction system in the mouse embryo heart. Dev. Biol. 80, 28–45 (1980).
Wang, J. et al. Pitx2 prevents susceptibility to atrial arrhythmias by inhibiting left-sided pacemaker specification. Proc. Natl Acad. Sci. USA 107, 9753–9758 (2010).
Ammirabile, G. et al. Pitx2 confers left morphological, molecular, and functional identity to the sinus venosus myocardium. Cardiovasc. Res. 93, 291–301 (2012).
Gudbjartsson, D. F. et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature 448, 353–357 (2007).
Wang, J. et al. Pitx2-microRNA pathway that delimits sinoatrial node development and inhibits predisposition to atrial fibrillation. Proc. Natl Acad. Sci. USA 111, 9181–9186 (2014).
Baudic, M. et al. TAD boundary deletion causes PITX2-related cardiac electrical and structural defects. Nat. Commun. 15, 3380 (2024).
Enard, W. et al. A humanized version of Foxp2 affects cortico-basal ganglia circuits in mice. Cell 137, 961–971 (2009).
Verweij, N., van de Vegte, Y. J. & van der Harst, P. Genetic study links components of the autonomous nervous system to heart-rate profile during exercise. Nat. Commun. 9, 898 (2018).
Bhattacharyya, S. et al. Global chromatin landscapes identify candidate noncoding modifiers of cardiac rhythm. J. Clin. Invest. 133, e153635 (2023).
van Eif, V. W. W. et al. Genome-wide analysis identifies an essential human TBX3 pacemaker enhancer. Circ. Res. 127, 1522–1535 (2020).
Galang, G. et al. ATAC-Seq reveals an Isl1 enhancer that regulates sinoatrial node development and function. Circ. Res. 127, 1502–1518 (2020).
Abassah-Oppong, S. et al. A gene desert required for regulatory control of pleiotropic Shox2 expression and embryonic survival. Nat. Commun. 15, 8793 (2024).
Ramirez, J. et al. Thirty loci identified for heart rate response to exercise and recovery implicate autonomic nervous system. Nat. Commun. 9, 1947 (2018).
Anderson, R. H., Mori, S., Spicer, D. E., Sanchez-Quintana, D. & Jensen, B. The anatomy, development, and evolution of the atrioventricular conduction axis. J. Cardiovasc. Dev. Dis. 5, 44 (2018).
de Carvalho, A. P. & de Almeida, D. F. Spread of activity through the atrioventricular node. Circ. Res. 8, 801–809 (1960).
Efimov, I. R. & Mazgalev, T. N. High-resolution, three-dimensional fluorescent imaging reveals multilayer conduction pattern in the atrioventricular node. Circulation 98, 54–57 (1998).
Aanhaanen, W. T. et al. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ. Res. 104, 1267–1274 (2009).
Horsthuis, T. et al. Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ. Res. 105, 61–69 (2009).
Anderson, R. H. et al. Inferior extensions of the atrioventricular node. Arrhythm. Electrophysiol. Rev. 10, 262–272 (2021).
Ma, L., Lu, M. F., Schwartz, R. J. & Martin, J. F. Bmp2 is essential for cardiac cushion epithelial–mesenchymal transition and myocardial patterning. Development 132, 5601–5611 (2005).
Chi, N. C. et al. Foxn4 directly regulates tbx2b expression and atrioventricular canal formation. Genes Dev. 22, 734–739 (2008).
Liu, F. et al. GATA-binding factor 6 contributes to atrioventricular node development and function. Circ. Cardiovasc. Genet. 8, 284–293 (2015).
Verhoeven, M. C., Haase, C., Christoffels, V. M., Weidinger, G. & Bakkers, J. Wnt signaling regulates atrioventricular canal formation upstream of BMP and Tbx2. Birth Defects Res. A Clin. Mol. Teratol. 91, 435–440 (2011).
Jay, P. Y. et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J. Clin. Invest. 113, 1130–1137 (2004).
Singh, R. et al. Tbx20 interacts with Smads to confine Tbx2 expression to the atrioventricular canal. Circ. Res. 105, 442–452 (2009).
Rutenberg, J. B. et al. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 133, 4381–4390 (2006).
Stefanovic, S. et al. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat. Commun. 5, 3680 (2014).
Singh, R. et al. Tbx2 and Tbx3 induce atrioventricular myocardial development and endocardial cushion formation. Cell Mol. Life Sci. 69, 1377–1389 (2012).
Bressan, M. et al. Reciprocal myocardial–endocardial interactions pattern the delay in atrioventricular junction conduction. Development 141, 4149–4157 (2014).
Lalani, S. R. et al. 20p12.3 microdeletion predisposes to Wolff–Parkinson–White syndrome with variable neurocognitive deficits. J. Med. Genet. 46, 168–175 (2009).
Gaussin, V. et al. Alk3/Bmpr1a receptor is required for development of the atrioventricular canal into valves and annulus fibrosus. Circ. Res. 97, 219–226 (2005).
Stroud, D. M. et al. Abnormal conduction and morphology in the atrioventricular node of mice with atrioventricular canal targeted deletion of Alk3/Bmpr1a receptor. Circulation 116, 2535–2543 (2007).
Aanhaanen, W. T. et al. Defective Tbx2-dependent patterning of the atrioventricular canal myocardium causes accessory pathway formation in mice. J. Clin. Invest. 121, 534–544 (2011).
Bakker, M. L. et al. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ. Res. 102, 1340–1349 (2008).
Pfeufer, A. et al. Genome-wide association study of PR interval. Nat. Genet. 42, 153–159 (2010).
Sotoodehnia, N. et al. Common variants in 22 loci are associated with QRS duration and cardiac ventricular conduction. Nat. Genet. 42, 1068–1076 (2010).
Verweij, N. et al. Genetic determinants of P wave duration and PR segment. Circ. Cardiovasc. Genet. 7, 475–481 (2014).
van Setten, J. et al. PR interval genome-wide association meta-analysis identifies 50 loci associated with atrial and atrioventricular electrical activity. Nat. Commun. 9, 2904 (2018).
Harrelson, Z. et al. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development 131, 5041–5052 (2004).
Smith, J. G. et al. Genome-wide association studies of the PR interval in African Americans. PLoS Genet. 7, e1001304 (2011).
Hong, K. W. et al. Identification of three novel genetic variations associated with electrocardiographic traits (QRS duration and PR interval) in East Asians. Hum. Mol. Genet. 23, 6659–6667 (2014).
Munshi, N. V. et al. Cx30.2 enhancer analysis identifies Gata4 as a novel regulator of atrioventricular delay. Development 136, 2665–2674 (2009).
Harris, J. P. et al. MyoR modulates cardiac conduction by repressing Gata4. Mol. Cell. Biol. 35, 649–661 (2015).
Pashmforoush, M. et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell 117, 373–386 (2004).
Ranganayakulu, G., Elliott, D. A., Harvey, R. P. & Olson, E. N. Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development 125, 3037–3048 (1998).
Tanaka, M., Chen, Z., Bartunkova, S., Yamasaki, N. & Izumo, S. The cardiac homeobox gene Csx/Nkx2.5 lies genetically upstream of multiple genes essential for heart development. Development 126, 1269–1280 (1999).
Munoz-Martin, N. et al. Meis transcription factors regulate cardiac conduction system development and adult function. Cardiovasc. Res. 121, 311–323 (2024).
Milan, D. J., Giokas, A. C., Serluca, F. C., Peterson, R. T. & MacRae, C. A. Notch1b and neuregulin are required for specification of central cardiac conduction tissue. Development 133, 1125–1132 (2006).
Rentschler, S. et al. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J. Clin. Invest. 121, 525–533 (2011).
Sun, T. et al. Dbh(+) catecholaminergic cardiomyocytes contribute to the structure and function of the cardiac conduction system in murine heart. Nat. Commun. 14, 7801 (2023).
Blackwell, D. J. et al. The Purkinje–myocardial junction is the anatomic origin of ventricular arrhythmia in CPVT. JCI Insight 7, e151893 (2022).
Gourdie, R. G., Mima, T., Thompson, R. P. & Mikawa, T. Terminal diversification of the myocyte lineage generates Purkinje fibers of the cardiac conduction system. Development 121, 1423–1431 (1995).
Rentschler, S. et al. Neuregulin-1 promotes formation of the murine cardiac conduction system. Proc. Natl Acad. Sci. USA 99, 10464–10469 (2002).
Choquet, C., Kelly, R. G. & Miquerol, L. Nkx2-5 defines distinct scaffold and recruitment phases during formation of the murine cardiac Purkinje fiber network. Nat. Commun. 11, 5300 (2020).
Choquet, C. et al. Nkx2-5 loss of function in the His-Purkinje system hampers its maturation and leads to mechanical dysfunction. J. Cardiovasc. Dev. Dis. 10, 194 (2023).
Moskowitz, I. P. et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell 129, 1365–1376 (2007).
Burnicka-Turek, O. et al. Transcriptional patterning of the ventricular cardiac conduction system. Circ. Res. 127, e94–e106 (2020).
Shekhar, A. et al. ETV1 activates a rapid conduction transcriptional program in rodent and human cardiomyocytes. Sci. Rep. 8, 9944 (2018).
Shekhar, A. et al. Transcription factor ETV1 is essential for rapid conduction in the heart. J. Clin. Invest. 126, 4444–4459 (2016).
Koizumi, A. et al. Genetic defects in a His-Purkinje system transcription factor, IRX3, cause lethal cardiac arrhythmias. Eur. Heart J. 37, 1469–1475 (2015).
Meysen, S. et al. Nkx2.5 cell-autonomous gene function is required for the postnatal formation of the peripheral ventricular conduction system. Dev. Biol. 303, 740–753 (2007).
Wakimoto, H., Kasahara, H., Maguire, C. T., Izumo, S. & Berul, C. I. Developmentally modulated cardiac conduction failure in transgenic mice with fetal or postnatal overexpression of DNA nonbinding mutant Nkx2.5. J. Cardiovasc. Electrophysiol. 13, 682–688 (2002).
Kasahara, H. et al. Progressive atrioventricular conduction defects and heart failure in mice expressing a mutant Csx/Nkx2.5 homeoprotein. J. Clin. Invest. 108, 189–201 (2001).
Risebro, C. A. et al. Epistatic rescue of Nkx2.5 adult cardiac conduction disease phenotypes by prospero-related homeobox protein 1 and HDAC3. Circ. Res. 111, e19–e31 (2012).
Yamaguchi, N. et al. An anterior second heart field enhancer regulates the gene regulatory network of the cardiac outflow tract. Circulation 148, 1705–1722 (2023).
Lien, C. L., McAnally, J., Richardson, J. A. & Olson, E. N. Cardiac-specific activity of an Nkx2-5 enhancer requires an evolutionarily conserved Smad binding site. Dev. Biol. 244, 257–266 (2002).
Lien, C. L. et al. Control of early cardiac-specific transcription of Nkx2-5 by a GATA-dependent enhancer. Development 126, 75–84 (1999).
Harris, B. S. et al. Differentiation of cardiac Purkinje fibers requires precise spatiotemporal regulation of Nkx2-5 expression. Dev. Dyn. 235, 38–49 (2006).
Arnolds, D. E. et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J. Clin. Invest. 122, 2509–2518 (2012).
Arnolds, D. E. & Moskowitz, I. P. Inducible recombination in the cardiac conduction system of minK: CreERT2 BAC transgenic mice. Genesis 49, 878–884 (2011).
Chambers, J. C. et al. Genetic variation in SCN10A influences cardiac conduction. Nat. Genet. 42, 149–152 (2010).
Holm, H. et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat. Genet. 42, 117–122 (2010).
Seyerle, A. A. et al. Genome-wide association study of PR interval in Hispanics/Latinos identifies novel locus at ID2. Heart 104, 904–911 (2018).
Christophersen, I. E. et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat. Genet. 49, 946–952 (2017).
van den Boogaard, M. et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J. Clin. Invest. 122, 2519–2530 (2012).
Man, J. C. K. et al. Variant intronic enhancer controls SCN10A-short expression and heart conduction. Circulation 144, 229–242 (2021).
van den Boogaard, M. et al. A common genetic variant within SCN10A modulates cardiac SCN5A expression. J. Clin. Invest. 124, 1844–1852 (2014).
Hua, L. L. et al. Specification of the mouse cardiac conduction system in the absence of Endothelin signaling. Dev. Biol. 393, 245–254 (2014).
Gourdie, R. G., Wei, Y., Kim, D., Klatt, S. C. & Mikawa, T. Endothelin-induced conversion of embryonic heart muscle cells into impulse-conducting Purkinje fibers. Proc. Natl Acad. Sci. USA 95, 6815–6818 (1998).
Patel, R. & Kos, L. Endothelin-1 and Neuregulin-1 convert embryonic cardiomyocytes into cells of the conduction system in the mouse. Dev. Dyn. 233, 20–28 (2005).
Yamaguchi, N. et al. Cardiac pressure overload decreases ETV1 expression in the left atrium, contributing to atrial electrical and structural remodeling. Circulation 143, 805–820 (2021).
Grego-Bessa, J. et al. Notch signaling is essential for ventricular chamber development. Dev. Cell 12, 415–429 (2007).
Ribeiro da Silva, A. et al. NOTCH1 is critical for fibroblast-mediated induction of cardiomyocyte specialization into ventricular conduction system-like cells in vitro. Sci. Rep. 10, 16163 (2020).
Gonzalez, D. M. et al. Transient notch activation converts pluripotent stem cell-derived cardiomyocytes towards a Purkinje fiber fate. Preprint at bioRxiv https://doi.org/10.1101/2024.09.22.614353 (2024).
Rentschler, S. et al. Myocardial Notch signaling reprograms cardiomyocytes to a conduction-like phenotype. Circulation 126, 1058–1066 (2012).
van Rijen, H. V. et al. Impaired conduction in the bundle branches of mouse hearts lacking the gap junction protein connexin40. Circulation 103, 1591–1598 (2001).
Tamaddon, H. S. et al. High-resolution optical mapping of the right bundle branch in connexin40 knockout mice reveals slow conduction in the specialized conduction system. Circ. Res. 87, 929–936 (2000).
Zhang, S. S. et al. Iroquois homeobox gene 3 establishes fast conduction in the cardiac His–Purkinje network. Proc. Natl Acad. Sci. USA 108, 13576–13581 (2011).
Kim, K. H. et al. Irx3 is required for postnatal maturation of the mouse ventricular conduction system. Sci. Rep. 6, 19197 (2016).
Chen, F. et al. Hop is an unusual homeobox gene that modulates cardiac development. Cell 110, 713–723 (2002).
Ismat, F. A. et al. Homeobox protein Hop functions in the adult cardiac conduction system. Circ. Res. 96, 898–903 (2005).
Evans, D. S. et al. Fine-mapping, novel loci identification, and SNP association transferability in a genome-wide association study of QRS duration in African Americans. Hum. Mol. Genet. 25, 4350–4368 (2016).
Vincentz, J. W. et al. Variation in a left ventricle-specific Hand1 enhancer impairs GATA transcription factor binding and disrupts conduction system development and function. Circ. Res. 125, 575–589 (2019).
Nguyen-Tran, V. T. et al. A novel genetic pathway for sudden cardiac death via defects in the transition between ventricular and conduction system cell lineages. Cell 102, 671–682 (2000).
Hewett, K. W. et al. Knockout of the neural and heart expressed gene HF-1b results in apical deficits of ventricular structure and activation. Cardiovasc. Res. 67, 548–560 (2005).
Umbarkar, P. et al. Cardiomyocyte SMAD4-dependent TGF-beta signaling is essential to maintain adult heart homeostasis. JACC Basic Transl. Sci. 4, 41–53 (2019).
Giri, B. et al. G4 resolvase 1 tightly binds and unwinds unimolecular G4-DNA. Nucleic Acids Res. 39, 7161–7178 (2011).
Gomez-Del Arco, P. et al. The G4 resolvase Dhx36 modulates cardiomyocyte differentiation and ventricular conduction system development. Nat. Commun. 15, 8602 (2024).
Haubner, B. J. et al. Functional recovery of a human neonatal heart after severe myocardial infarction. Circ. Res. 118, 216–221 (2016).
Porrello, E. R. et al. Transient regenerative potential of the neonatal mouse heart. Science 331, 1078–1080 (2011).
Wang, H. et al. Electrophysiologic conservation of epicardial conduction dynamics after myocardial infarction and natural heart regeneration in newborn piglets. Front. Cardiovasc. Med. 9, 829546 (2022).
Wang, L., Bhakta, M., Fernandez-Perez, A. & Munshi, N. V. Inducible cardiomyocyte injury within the atrioventricular conduction system uncovers latent regenerative capacity in mice. J. Clin. Invest. 131, e138637 (2021).
Kahr, P. C. et al. A novel transgenic Cre allele to label mouse cardiac conduction system. Dev. Biol. 478, 163–172 (2021).
Boulgakoff, L. et al. Participation of ventricular trabeculae in neonatal cardiac regeneration leads to ectopic recruitment of Purkinje-like cells. Nat. Cardiovasc. Res. 3, 1140–1157 (2024).
Sayers, J. R. et al. Cardiac conduction system regeneration prevents arrhythmias after myocardial infarction. Nat. Cardiovasc. Res. 4, 163–179 (2025).
Meyer, D. & Birchmeier, C. Multiple essential functions of neuregulin in development. Nature 378, 386–390 (1995).
Lai, D. et al. Neuregulin 1 sustains the gene regulatory network in both trabecular and nontrabecular myocardium. Circ. Res. 107, 715–727 (2010).
Kim, E. E. et al. The transcription factor EBF1 non-cell-autonomously regulates cardiac growth and differentiation. Development 150, dev202054 (2023).
Park, D. S. et al. Pocket proteins critically regulate cell cycle exit of the trabecular myocardium and the ventricular conduction system. Biol. Open 2, 968–978 (2013).
Csepe, T. A., Kalyanasundaram, A., Hansen, B. J., Zhao, J. & Fedorov, V. V. Fibrosis: a structural modulator of sinoatrial node physiology and dysfunction. Front. Physiol. 6, 37 (2015).
Gluck, J. M. et al. Biochemical and biomechanical properties of the pacemaking sinoatrial node extracellular matrix are distinct from contractile left ventricular matrix. PLoS ONE 12, e0185125 (2017).
Bressan, M. et al. Dynamic cellular integration drives functional assembly of the heart’s pacemaker complex. Cell Rep. 23, 2283–2291 (2018).
Sun, Y. H. et al. The sinoatrial node extracellular matrix promotes pacemaker phenotype and protects automaticity in engineered heart tissues from cyclic strain. Cell Rep. 42, 113505 (2023).
Kalyanasundaram, A. et al. Fibroblast-specific proteotranscriptomes reveal distinct fibrotic signatures of human sinoatrial node in nonfailing and failing hearts. Circulation 144, 126–143 (2021).
Restrepo, A. J., Razminia, M., Sanchez-Quintana, D. & Cabrera, J. A. Myoarchitecture of the sinoatrial node and its relevance for catheter ablation: anatomy and histology. JACC Case Rep. 29, 102153 (2024).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Sawa, H. et al. Expression of the angiotensinogen gene and localization of its protein in the human heart. Circulation 86, 138–146 (1992).
Ieda, M. et al. Sema3a maintains normal heart rhythm through sympathetic innervation patterning. Nat. Med. 13, 604–612 (2007).
Hulsmans, M. et al. Macrophages facilitate electrical conduction in the heart. Cell 169, 510–522.e20 (2017).
Roselli, C. et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat. Genet. 50, 1225–1233 (2018).
Nielsen, J. B. et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat. Genet. 50, 1234–1239 (2018).
Li, J. et al. Modeling the atrioventricular conduction axis using human pluripotent stem cell-derived cardiac assembloids. Cell Stem Cell 31, 1667–1684.e6 (2024).
Qu, J. et al. Expression and function of a biological pacemaker in canine heart. Circulation 107, 1106–1109 (2003).
Miake, J., Marban, E. & Nuss, H. B. Biological pacemaker created by gene transfer. Nature 419, 132–133 (2002).
Boink, G. J. et al. HCN2/SkM1 gene transfer into canine left bundle branch induces stable, autonomically responsive biological pacing at physiological heart rates. J. Am. Coll. Cardiol. 61, 1192–1201 (2013).
Cingolani, E. et al. Biological pacemaker created by percutaneous gene delivery via venous catheters in a porcine model of complete heart block. Heart Rhythm 9, 1310–1318 (2012).
Hu, Y. F., Dawkins, J. F., Cho, H. C., Marban, E. & Cingolani, E. Biological pacemaker created by minimally invasive somatic reprogramming in pigs with complete heart block. Sci. Transl. Med. 6, 245ra294 (2014).
Fan, J. et al. Inhibition of Tgf-beta signaling enables durable ventricular pacing by TBX18 gene transfer. Preprint at bioRxiv https://doi.org/10.1101/2022.06.02.493572 (2022).
Keith, A. & Flack, M. The form and nature of the muscular connections between the primary divisions of the vertebrate heart. J. Anat. Physiol. 41, 172–189 (1907).
His, W. Jr. The story of the atrioventricular bundle with remarks concerning embryonic heart activity. J. Hist. Med. Allied Sci. 4, 319–333 (1949).
Suma, K. Sunao Tawara: a father of modern cardiology. Pacing Clin. Electrophysiol. 24, 88–96 (2001).
Steiner, I. History of the Purkyne fibres in the heart. On the occasion of bicentenary of Jan Evangelista Purkyne’s birth (December 17, 1787). Sb. Ved. Pr. Lek. Fak. Karlov. Univerzity Hradci Kralove 30, 7–20 (1987).
Roselli, C., Rienstra, M. & Ellinor, P. T. Genetics of atrial fibrillation in 2020: GWAS, genome sequencing, polygenic risk, and beyond. Circ. Res. 127, 21–33 (2020).
Koizumi, A. et al. Genetic defects in a His–Purkinje system transcription factor, IRX3, cause lethal cardiac arrhythmias. Eur. Heart J. 37, 1469–1475 (2016).
Tessadori, F. et al. Identification and functional characterization of cardiac pacemaker cells in zebrafish. PLoS ONE 7, e47644 (2012).
Naya, F. J. et al. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 8, 1303–1309 (2002).
Ritchie, M. D. et al. Genome- and phenome-wide analyses of cardiac conduction identifies markers of arrhythmia risk. Circulation 127, 1377–1385 (2013).
Benson, D. W. et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. J. Clin. Invest. 104, 1567–1573 (1999).
Costa, M. W. et al. Functional characterization of a novel mutation in NKX2-5 associated with congenital heart disease and adult-onset cardiomyopathy. Circ. Cardiovasc. Genet. 6, 238–247 (2013).
Yuan, F. et al. A novel NKX2-5 loss-of-function mutation predisposes to familial dilated cardiomyopathy and arrhythmias. Int. J. Mol. Med. 35, 478–486 (2015).
den Hoed, M. et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat. Genet. 45, 621–631 (2013).
Takeda, M. et al. Slow progressive conduction and contraction defects in loss of Nkx2-5 mice after cardiomyocyte terminal differentiation. Lab. Invest. 89, 983–993 (2009).
Kääb, S. et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur. Heart J. 30, 813–819 (2009).
Chinchilla, A. et al. PITX2 insufficiency leads to atrial electrical and structural remodeling linked to arrhythmogenesis. Circ. Cardiovasc. Genet. 4, 269–279 (2011).
Tao, Y. et al. Pitx2, an atrial fibrillation predisposition gene, directly regulates ion transport and intercalated disc genes. Circ. Cardiovasc. Genet. 7, 23–32 (2014).
Li, N. et al. A SHOX2 loss-of-function mutation underlying familial atrial fibrillation. Int. J. Med. Sci. 15, 1564–1572 (2018).
Hoffmann, S. et al. Coding and non-coding variants in the SHOX2 gene in patients with early-onset atrial fibrillation. Basic Res. Cardiol. 111, 36 (2016).
Umbarkar, P. et al. Cardiomyocyte SMAD4-dependent TGF-β signaling is essential to maintain adult heart homeostasis. JACC Basic Transl. Sci. 4, 41–53 (2019).
Tegegne, B. S. et al. Phenotypic but not genetically predicted heart rate variability associated with all-cause mortality. Commun. Biol. 6, 1013 (2023).
Nolte, I. M. et al. Genetic loci associated with heart rate variability and their effects on cardiac disease risk. Nat. Commun. 8, 15805 (2017).
Sano, M. et al. Genome-wide association study of absolute QRS voltage identifies common variants of TBX3 as genetic determinants of left ventricular mass in a healthy Japanese population. PLoS ONE 11, e0155550 (2016).
van der Harst, P. et al. 52 Genetic loci influencing myocardial mass. J. Am. Coll. Cardiol. 68, 1435–1448 (2016).
Jameson, H. S. et al. Loss of the atrial fibrillation-related gene, Zfhx3, results in atrial dilation and arrhythmias. Circ. Res. 133, 313–329 (2023).
Hill, M. C. et al. A cellular atlas of Pitx2-dependent cardiac development. Development 146, dev180398 (2019).
Ren, H. et al. Single-cell RNA sequencing of murine hearts for studying the development of the cardiac conduction system. Sci. Data 10, 577 (2023).
Feulner, L. et al. Transcriptional dynamics of the murine heart during perinatal development at single-cell resolution. Preprint at bioRxiv https://doi.org/10.1101/2024.03.05.583423 (2024).
Park, D. S. & Fishman, G. I. in Cardiac Electrophysiology: From Cell to Bedside (eds Zipes, D. P. & Jalife, J.) 287–295 (Elsevier, 2018).
Acknowledgements
D.S.P. receives research funding from the National Institutes of Health (R01HL165130 and R01HL171989). G.I.F. receives research funding from the National Institutes of Health (R01 HL171118 and R01HL159869). The authors thank N. Yamaguchi (NYU Grossman School of Medicine, USA) for assistance with the generation of tables and figures for initial submission.
Author information
Authors and Affiliations
Contributions
The authors contributed substantially to all aspects of the article.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Cardiology thanks Nikhil V. Munshi, Kalyanam Shivkumar and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Park, D.S., Fishman, G.I. The cardiac conduction system: development, function and therapeutic targets. Nat Rev Cardiol (2026). https://doi.org/10.1038/s41569-025-01227-x
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41569-025-01227-x