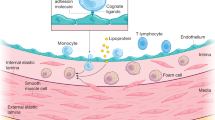
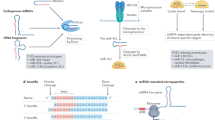
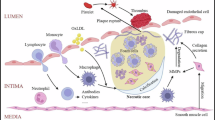
Abstract
The discovery of non-coding RNAs has expanded our understanding of how genetic features are linked to cellular function. The illumination of this so-called dark matter of the genome has revealed new categories of RNA with essential roles in the regulation of protein-coding genes and genome organization. In particular, microRNAs and long non-coding RNAs have emerged as important regulators of cardiovascular health and disease. In this Review, we summarize our current understanding of the mechanisms and functional roles of microRNAs and long non-coding RNAs in the regulation of lipid homeostasis, vascular biology and atherosclerosis. We discuss how interruption of non-coding RNA regulatory circuits influence lipoprotein metabolism in the liver and the circulation, as well as the effects of non-coding RNAs on inflammatory processes in the artery wall that contribute to atherosclerotic plaque formation. Finally, we highlight potential opportunities to harness non-coding RNAs as biomarkers and targeted therapeutics for atherosclerotic cardiovascular disease.
Key points
-
The discovery and characterization of non-coding RNAs have upended our view of the genome, revealing a vast RNA regulatory network that orchestrates cellular function beyond protein coding.
-
MicroRNAs — small (~22 nucleotides) non-coding RNAs that guide Argonaute complexes to target mRNAs for translational repression or degradation — fine-tune gene networks governing lipoprotein synthesis, lipoprotein clearance and reverse cholesterol transport, as well as cellular processes in atherosclerosis.
-
Long non-coding RNAs — transcripts of >200 nucleotides that do not code for proteins — are versatile regulators of genome organization and gene expression that act by scaffolding chromatin modifiers, guiding transcriptional complexes, modulating RNA splicing or stability and sequestering regulatory molecules.
-
Long non-coding RNAs have roles in lipid metabolism and atherosclerosis by coordinating the transcriptional and post-transcriptional regulation of cholesterol biosynthesis and efflux, triglyceride homeostasis and inflammatory and phenotypic programmes in vascular and immune cells.
-
Extracellular RNA communication via extracellular vesicles, HDL particles and Argonaute-bound complexes reveals intercellular signalling pathways, organ-to-organ regulatory circuits and novel biomarkers in cardiovascular disease.
-
Therapeutic manipulation of microRNAs and long non-coding RNAs holds transformative potential for atherosclerotic cardiovascular disease, but its clinical adoption requires overcoming barriers in delivery, targeting precision and immunogenicity.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Orgel, L. E. & Crick, F. H. Selfish DNA: the ultimate parasite. Nature 284, 604–607 (1980).
Mattick, J. S. et al. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447 (2023).
Feinberg, M. W. & Moore, K. J. MicroRNA regulation of atherosclerosis. Circ. Res. 118, 703–720 (2016).
Zhang, Z., Salisbury, D. & Sallam, T. Long noncoding RNAs in atherosclerosis: JACC review topic of the week. J. Am. Coll. Cardiol. 72, 2380–2390 (2018).
Ference, B. A. et al. Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement from the European Atherosclerosis Society Consensus Panel. Eur. Heart J. 38, 2459–2472 (2017).
Nordestgaard, B. G. Triglyceride-rich lipoproteins and atherosclerotic cardiovascular disease: new insights from epidemiology, genetics, and biology. Circ. Res. 118, 547–563 (2016).
Bonaccorsi, I. et al. Natural killer cells in the innate immunity network of atherosclerosis. Immunol. Lett. 168, 51–57 (2015).
Bjorkegren, J. L. M. & Lusis, A. J. Atherosclerosis: recent developments. Cell 185, 1630–1645 (2022).
Bennett, M. R., Sinha, S. & Owens, G. K. Vascular smooth muscle cells in atherosclerosis. Circ. Res. 118, 692–702 (2016).
Hansson, G. K. & Libby, P. The immune response in atherosclerosis: a double-edged sword. Nat. Rev. Immunol. 6, 508–519 (2006).
Fernandez-Hernando, C., Suarez, Y., Rayner, K. J. & Moore, K. J. MicroRNAs in lipid metabolism. Curr. Opin. Lipidol. 22, 86–92 (2011).
Moore, K. J., Rayner, K. J., Suarez, Y. & Fernandez-Hernando, C. MicroRNAs and cholesterol metabolism. Trends Endocrinol. Metab. 21, 699–706 (2010).
Urbich, C., Kuehbacher, A. & Dimmeler, S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc. Res. 79, 581–588 (2008).
Bink, D. I., Pauli, J., Maegdefessel, L. & Boon, R. A. Endothelial microRNAs and long noncoding RNAs in cardiovascular ageing. Atherosclerosis 374, 99–106 (2023).
Brown, S. D., Klimi, E., Bakker, W. A. M., Beqqali, A. & Baker, A. H. Non-coding RNAs to treat vascular smooth muscle cell dysfunction. Br. J. Pharmacol. 182, 246–280 (2025).
Sekine, S. et al. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology 136, 2304–2315.e4 (2009).
Lagos-Quintana, M. et al. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 12, 735–739 (2002).
Elmen, J. et al. LNA-mediated microRNA silencing in non-human primates. Nature 452, 896–899 (2008).
Esau, C. et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3, 87–98 (2006).
Doddapaneni, R. et al. Overexpression of microRNA-122 enhances in vitro hepatic differentiation of fetal liver-derived stem/progenitor cells. J. Cell. Biochem. 114, 1575–1583 (2013).
Krutzfeldt, J. et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 438, 685–689 (2005).
Girard, M., Jacquemin, E., Munnich, A., Lyonnet, S. & Henrion-Caude, A. miR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 48, 648–656 (2008).
Soh, J., Iqbal, J., Queiroz, J., Fernandez-Hernando, C. & Hussain, M. M. MicroRNA-30c reduces hyperlipidemia and atherosclerosis in mice by decreasing lipid synthesis and lipoprotein secretion. Nat. Med. 19, 892–900 (2013).
Irani, S. et al. MicroRNA-30c mimic mitigates hypercholesterolemia and atherosclerosis in mice. J. Biol. Chem. 291, 18397–18409 (2016).
Zhang, M., Sun, W., Zhou, M. & Tang, Y. MicroRNA-27a regulates hepatic lipid metabolism and alleviates NAFLD via repressing FAS and SCD1. Sci. Rep. 7, 14493 (2017).
Ji, J. et al. Over-expressed microRNA-27a and 27b influence fat accumulation and cell proliferation during rat hepatic stellate cell activation. FEBS Lett. 583, 759–766 (2009).
Vickers, K. C. et al. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 57, 533–542 (2013).
Wagschal, A. et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 21, 1290–1297 (2015).
Sakai, E. et al. miR-27b targets MAIP1 to mediate lipid accumulation in cultured human and mouse hepatic cells. Commun. Biol. 6, 669 (2023).
Tan, Y., Ge, G., Pan, T., Wen, D. & Gan, J. A pilot study of serum microRNAs panel as potential biomarkers for diagnosis of nonalcoholic fatty liver disease. PLoS ONE 9, e105192 (2014).
Ando, Y. et al. Association of circulating miR-20a, miR-27a, and miR-126 with non-alcoholic fatty liver disease in general population. Sci. Rep. 9, 18856 (2019).
Goedeke, L. et al. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis 243, 499–509 (2015).
Tang, Y. et al. Exosomal miR-27b-3p secreted by visceral adipocytes contributes to endothelial inflammation and atherogenesis. Cell Rep. 42, 111948 (2023).
Vickers, K. C. et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl Acad. Sci. USA 111, 14518–14523 (2014).
Singaravelu, R. et al. MicroRNA-7 mediates cross-talk between metabolic signaling pathways in the liver. Sci. Rep. 8, 361 (2018).
Xu, Y. et al. A metabolic stress-inducible miR-34a-HNF4alpha pathway regulates lipid and lipoprotein metabolism. Nat. Commun. 6, 7466 (2015).
Ding, J. et al. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Sci. Rep. 5, 13729 (2015).
Goedeke, L. et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 21, 1280–1289 (2015).
Jiang, H. et al. microRNA-185 modulates low density lipoprotein receptor expression as a key posttranscriptional regulator. Atherosclerosis 243, 523–532 (2015).
Aranda, J. F., Canfran-Duque, A., Goedeke, L., Suarez, Y. & Fernandez-Hernando, C. The miR-199-dynamin regulatory axis controls receptor-mediated endocytosis. J. Cell Sci. 128, 3197–3209 (2015).
Salerno, A. G. et al. LDL receptor pathway regulation by miR-224 and miR-520d. Front. Cardiovasc. Med. 7, 81 (2020).
van Solingen, C., Oldebeken, S. R., Salerno, A. G., Wanschel, A. & Moore, K. J. High-throughput screening identifies microRNAs regulating human PCSK9 and hepatic low-density lipoprotein receptor expression. Front. Cardiovasc. Med. 8, 667298 (2021).
Dong, J. et al. microRNA-483 ameliorates hypercholesterolemia by inhibiting PCSK9 production. JCI Insight 5, e143812 (2020).
Wang, Y. et al. miR-181d-5p ameliorates hypercholesterolemia by targeting PCSK9. J. Endocrinol. 262, e230402 (2024).
Xu, X. et al. MiR-337-3p lowers serum LDL-C level through targeting PCSK9 in hyperlipidemic mice. Metabolism 119, 154768 (2021).
Chen, X. et al. MiR-99a-5p up-regulates LDLR and functionally enhances LDL-C uptake via suppressing PCSK9 expression in human hepatocytes. Front. Genet. 15, 1469094 (2024).
Ma, N. et al. New PCSK9 inhibitor miR-552-3p reduces LDL-C via enhancing LDLR in high fat diet-fed mice. Pharmacol. Res. 167, 105562 (2021).
Rayner, K. J. et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature 478, 404–407 (2011).
Rayner, K. J. et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Invest. 121, 2921–2931 (2011).
Rayner, K. J. et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 328, 1570–1573 (2010).
Marquart, T. J., Allen, R. M., Ory, D. S. & Baldan, A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl Acad. Sci. USA 107, 12228–12232 (2010).
Najafi-Shoushtari, S. H. et al. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 328, 1566–1569 (2010).
Ramirez, C. M. et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ. Res. 112, 1592–1601 (2013).
de Aguiar Vallim, T. Q. et al. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 112, 1602–1612 (2013).
Gerin, I. et al. Expression of miR-33 from an SREBP2 intron inhibits cholesterol export and fatty acid oxidation. J. Biol. Chem. 285, 33652–33661 (2010).
Davalos, A. et al. miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc. Natl Acad. Sci. USA 108, 9232–9237 (2011).
Rottiers, V. et al. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci. Transl. Med. 5, 212ra162 (2013).
Afonso, M. S. et al. miR-33 silencing reprograms the immune cell landscape in atherosclerotic plaques. Circ. Res. 128, 1122–1138 (2021).
Ouimet, M. et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Invest. 125, 4334–4348 (2015).
Rotllan, N., Ramirez, C. M., Aryal, B., Esau, C. C. & Fernandez-Hernando, C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr−/− mice — brief report. Arterioscler. Thromb. Vasc. Biol. 33, 1973–1977 (2013).
Price, N. L. et al. Genetic dissection of the impact of miR-33a and miR-33b during the progression of atherosclerosis. Cell Rep. 21, 1317–1330 (2017).
Nishino, T. et al. SREBF1/microRNA-33b axis exhibits potent effect on unstable atherosclerotic plaque formation in vivo. Arterioscler. Thromb. Vasc. Biol. 38, 2460–2473 (2018).
Price, N. L. et al. Genetic ablation of miR-33 increases food intake, enhances adipose tissue expansion, and promotes obesity and insulin resistance. Cell Rep. 22, 2133–2145 (2018).
Goedeke, L. et al. Long-term therapeutic silencing of miR-33 increases circulating triglyceride levels and hepatic lipid accumulation in mice. EMBO Mol. Med. 6, 1133–1141 (2014).
Horie, T. et al. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat. Commun. 4, 2883 (2013).
Cheng, J. et al. MicroRNA-144 silencing protects against atherosclerosis in male, but not female mice. Arterioscler. Thromb. Vasc. Biol. 40, 412–425 (2020).
Xu, Y. et al. Macrophage miR-34a is a key regulator of cholesterol efflux and atherosclerosis. Mol. Ther. 28, 202–216 (2020).
Canfran-Duque, A., Lin, C. S., Goedeke, L., Suarez, Y. & Fernandez-Hernando, C. Micro-RNAs and high-density lipoprotein metabolism. Arterioscler. Thromb. Vasc. Biol. 36, 1076–1084 (2016).
Hartmann, P. et al. Endothelial Dicer promotes atherosclerosis and vascular inflammation by miRNA-103-mediated suppression of KLF4. Nat. Commun. 7, 10521 (2016).
Joshi, D. et al. Endothelial gamma-protocadherins inhibit KLF2 and KLF4 to promote atherosclerosis. Nat. Cardiovasc. Res. 3, 1035–1048 (2024).
Fang, Y. & Davies, P. F. Site-specific microRNA-92a regulation of Krüppel-like factors 4 and 2 in atherosusceptible endothelium. Arterioscler. Thromb. Vasc. Biol. 32, 979–987 (2012).
Loyer, X. et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ. Res. 114, 434–443 (2014).
Chang, Y. J. et al. Extracellular microRNA-92a mediates endothelial cell–macrophage communication. Arterioscler. Thromb. Vasc. Biol. 39, 2492–2504 (2019).
Liu, Y. et al. Atherosclerotic conditions promote the packaging of functional microRNA-92a-3p into endothelial microvesicles. Circ. Res. 124, 575–587 (2019).
Shang, F. et al. MicroRNA-92a mediates endothelial dysfunction in CKD. J. Am. Soc. Nephrol. 28, 3251–3261 (2017).
Huang, Y. et al. Circulating miR-92a expression level in patients with essential hypertension: a potential marker of atherosclerosis. J. Hum. Hypertens. 31, 200–205 (2017).
Abplanalp, W. T. et al. Efficiency and target derepression of anti-miR-92a: results of a first in human study. Nucleic Acid. Ther. 30, 335–345 (2020).
Daniel, J. M. et al. Inhibition of miR-92a improves re-endothelialization and prevents neointima formation following vascular injury. Cardiovasc. Res. 103, 564–572 (2014).
Zhou, Z. et al. Targeted polyelectrolyte complex micelles treat vascular complications in vivo. Proc. Natl Acad. Sci. USA 118, e2114842118 (2021).
Fish, J. E. et al. miR-126 regulates angiogenic signaling and vascular integrity. Dev. Cell 15, 272–284 (2008).
Wang, S. et al. The endothelial-specific microRNA miR-126 governs vascular integrity and angiogenesis. Dev. Cell 15, 261–271 (2008).
van Solingen, C. et al. Antagomir-mediated silencing of endothelial cell specific microRNA-126 impairs ischemia-induced angiogenesis. J. Cell. Mol. Med. 13, 1577–1585 (2009).
Schober, A. et al. MicroRNA-126-5p promotes endothelial proliferation and limits atherosclerosis by suppressing Dlk1. Nat. Med. 20, 368–376 (2014).
Santovito, D. et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci. Transl. Med. 12, eaaz2294 (2020).
Harris, T. A., Yamakuchi, M., Ferlito, M., Mendell, J. T. & Lowenstein, C. J. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl Acad. Sci. USA 105, 1516–1521 (2008).
Asgeirsdottir, S. A. et al. MicroRNA-126 contributes to renal microvascular heterogeneity of VCAM-1 protein expression in acute inflammation. Am. J. Physiol. Ren. Physiol. 302, F1630–F1639 (2012).
Jansen, F. et al. Intercellular transfer of miR-126-3p by endothelial microparticles reduces vascular smooth muscle cell proliferation and limits neointima formation by inhibiting LRP6. J. Mol. Cell. Cardiol. 104, 43–52 (2017).
Zernecke, A. et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci. Signal. 2, ra81 (2009).
van Solingen, C. et al. MicroRNA-126 modulates endothelial SDF-1 expression and mobilization of Sca-1+/Lin− progenitor cells in ischaemia. Cardiovasc. Res. 92, 449–455 (2011).
Lv, B. et al. MicroRNA-181 in cardiovascular disease: emerging biomarkers and therapeutic targets. FASEB J. 38, e23635 (2024).
Su, Y. et al. MicroRNA-181a-5p and microRNA-181a-3p cooperatively restrict vascular inflammation and atherosclerosis. Cell Death Dis. 10, 365 (2019).
Lin, J. et al. MicroRNA-181b inhibits thrombin-mediated endothelial activation and arterial thrombosis by targeting caspase recruitment domain family member 10. FASEB J. 30, 3216–3226 (2016).
Sun, X. et al. Systemic delivery of microRNA-181b inhibits nuclear factor-kappaB activation, vascular inflammation, and atherosclerosis in apolipoprotein E-deficient mice. Circ. Res. 114, 32–40 (2014).
Fernandez-Hernando, C. & Suarez, Y. MicroRNAs in endothelial cell homeostasis and vascular disease. Curr. Opin. Hematol. 25, 227–236 (2018).
Lu, L. et al. Time series miRNA–mRNA integrated analysis reveals critical miRNAs and targets in macrophage polarization. Sci. Rep. 6, 37446 (2016).
Zhang, Y., Zhang, M., Zhong, M., Suo, Q. & Lv, K. Expression profiles of miRNAs in polarized macrophages. Int. J. Mol. Med. 31, 797–802 (2013).
Baer, C. et al. Suppression of microRNA activity amplifies IFN-gamma-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol. 18, 790–802 (2016).
Wei, Y. et al. Dicer in macrophages prevents atherosclerosis by promoting mitochondrial oxidative metabolism. Circulation 138, 2007–2020 (2018).
Du, F. et al. MicroRNA-155 deficiency results in decreased macrophage inflammation and attenuated atherogenesis in apolipoprotein E-deficient mice. Arterioscler. Thromb. Vasc. Biol. 34, 759–767 (2014).
Nazari-Jahantigh, M. et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. J. Clin. Invest. 122, 4190–4202 (2012).
Wei, Y. et al. The microRNA-342-5p fosters inflammatory macrophage activation through an Akt1- and microRNA-155-dependent pathway during atherosclerosis. Circulation 127, 1609–1619 (2013).
Androulidaki, A. et al. The kinase Akt1 controls macrophage response to lipopolysaccharide by regulating microRNAs. Immunity 31, 220–231 (2009).
Mann, M. et al. An NF-kappaB-microRNA regulatory network tunes macrophage inflammatory responses. Nat. Commun. 8, 851 (2017).
O’Connell, R. M., Chaudhuri, A. A., Rao, D. S. & Baltimore, D. Inositol phosphatase SHIP1 is a primary target of miR-155. Proc. Natl Acad. Sci. USA 106, 7113–7118 (2009).
O’Connell, R. M., Taganov, K. D., Boldin, M. P., Cheng, G. & Baltimore, D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc. Natl Acad. Sci. USA 104, 1604–1609 (2007).
van Solingen, C., Araldi, E., Chamorro-Jorganes, A., Fernandez-Hernando, C. & Suarez, Y. Improved repair of dermal wounds in mice lacking microRNA-155. J. Cell. Mol. Med. 18, 1104–1112 (2014).
Curtis, A. M. et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Natl Acad. Sci. USA 112, 7231–7236 (2015).
Donners, M. M. et al. Hematopoietic miR155 deficiency enhances atherosclerosis and decreases plaque stability in hyperlipidemic mice. PLoS ONE 7, e35877 (2012).
Hou, J. et al. MicroRNA-146a feedback inhibits RIG-I-dependent type I IFN production in macrophages by targeting TRAF6, IRAK1, and IRAK2. J. Immunol. 183, 2150–2158 (2009).
Taganov, K. D., Boldin, M. P., Chang, K. J. & Baltimore, D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA 103, 12481–12486 (2006).
Raitoharju, E. et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 219, 211–217 (2011).
Sheedy, F. J. et al. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat. Immunol. 11, 141–147 (2010).
Das, A., Ganesh, K., Khanna, S., Sen, C. K. & Roy, S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J. Immunol. 192, 1120–1129 (2014).
Canfran-Duque, A. et al. Macrophage deficiency of miR-21 promotes apoptosis, plaque necrosis, and vascular inflammation during atherogenesis. EMBO Mol. Med. 9, 1244–1262 (2017).
Fan, X., Wang, E., Wang, X., Cong, X. & Chen, X. MicroRNA-21 is a unique signature associated with coronary plaque instability in humans by regulating matrix metalloproteinase-9 via reversion-inducing cysteine-rich protein with Kazal motifs. Exp. Mol. Pathol. 96, 242–249 (2014).
Hoekstra, M. et al. The peripheral blood mononuclear cell microRNA signature of coronary artery disease. Biochem. Biophys. Res. Commun. 394, 792–797 (2010).
Xi, J. et al. miR-21 depletion in macrophages promotes tumoricidal polarization and enhances PD-1 immunotherapy. Oncogene 37, 3151–3165 (2018).
Hackett, E. E. et al. Mycobacterium tuberculosis limits host glycolysis and IL-1beta by restriction of PFK-M via MicroRNA-21. Cell Rep. 30, 124–136.e4 (2020).
Wang, D. et al. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ. Res. 111, 967–981 (2012).
Sun, D. et al. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 586, 1472–1479 (2012).
Zhang, N. et al. MicroRNA-101 overexpression by IL-6 and TNF-alpha inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression. Exp. Cell Res. 336, 33–42 (2015).
Meiler, S., Baumer, Y., Toulmin, E., Seng, K. & Boisvert, W. A. MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 35, 323–331 (2015).
Ramirez, C. M. et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 31, 2707–2714 (2011).
Price, N. L., Goedeke, L., Suarez, Y. & Fernandez-Hernando, C. miR-33 in cardiometabolic diseases: lessons learned from novel animal models and approaches. EMBO Mol. Med. 13, e12606 (2021).
Ouimet, M. et al. MicroRNA-33 regulates macrophage autophagy in atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 37, 1058–1067 (2017).
Ouimet, M. et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 17, 677–686 (2016).
Ouimet, M. et al. miRNA targeting of oxysterol-binding protein-like 6 regulates cholesterol trafficking and efflux. Arterioscler. Thromb. Vasc. Biol. 36, 942–951 (2016).
Price, N. L. et al. Loss of hepatic miR-33 improves metabolic homeostasis and liver function without altering body weight or atherosclerosis. Proc. Natl Acad. Sci. USA 118, e2006478118 (2021).
Rahman, K. et al. Inflammatory Ly6Chi monocytes and their conversion to M2 macrophages drive atherosclerosis regression. J. Clin. Invest. 127, 2904–2915 (2017).
Ying, W. et al. MicroRNA-223 is a crucial mediator of PPARgamma-regulated alternative macrophage activation. J. Clin. Invest. 125, 4149–4159 (2015).
Zhuang, G. et al. A novel regulator of macrophage activation: miR-223 in obesity-associated adipose tissue inflammation. Circulation 125, 2892–2903 (2012).
Nguyen, M. A. et al. miR-223 exerts translational control of proatherogenic genes in macrophages. Circ. Res. 131, 42–58 (2022).
Jia, Y. et al. miR-223-3p prevents necroptotic macrophage death by targeting ripk3 in a negative feedback loop and consequently ameliorates advanced atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 44, 218–237 (2024).
McCubbrey, A. L. et al. MicroRNA-34a negatively regulates efferocytosis by tissue macrophages in part via SIRT1. J. Immunol. 196, 1366–1375 (2016).
Basatemur, G. L., Jorgensen, H. F., Clarke, M. C. H., Bennett, M. R. & Mallat, Z. Vascular smooth muscle cells in atherosclerosis. Nat. Rev. Cardiol. 16, 727–744 (2019).
Chen, R., McVey, D. G., Shen, D., Huang, X. & Ye, S. Phenotypic switching of vascular smooth muscle cells in atherosclerosis. J. Am. Heart Assoc. 12, e031121 (2023).
Jang, B. et al. MicroRNAs in vascular smooth muscle cells: mechanisms, therapeutic potential, and advances in delivery systems. Life Sci. 364, 123424 (2025).
Vacante, F., Denby, L., Sluimer, J. C. & Baker, A. H. The function of miR-143, miR-145 and the MiR-143 host gene in cardiovascular development and disease. Vasc. Pharmacol. 112, 24–30 (2019).
Lovren, F. et al. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation 126, S81–S90 (2012).
Climent, M. et al. TGFbeta triggers miR-143/145 transfer from smooth muscle cells to endothelial cells, thereby modulating vessel stabilization. Circ. Res. 116, 1753–1764 (2015).
Leeper, N. J. & Maegdefessel, L. Non-coding RNAs: key regulators of smooth muscle cell fate in vascular disease. Cardiovasc. Res. 114, 611–621 (2018).
Farina, F. M. et al. miR-128-3p is a novel regulator of vascular smooth muscle cell phenotypic switch and vascular diseases. Circ. Res. 126, e120–e135 (2020).
Zhang, J. et al. High-content analysis of microRNAs involved in the phenotype regulation of vascular smooth muscle cells. Sci. Rep. 12, 3498 (2022).
Durham, A. L., Speer, M. Y., Scatena, M., Giachelli, C. M. & Shanahan, C. M. Role of smooth muscle cells in vascular calcification: implications in atherosclerosis and arterial stiffness. Cardiovasc. Res. 114, 590–600 (2018).
Liao, X. B. et al. MiR-133a modulates osteogenic differentiation of vascular smooth muscle cells. Endocrinology 154, 3344–3352 (2013).
Goettsch, C. et al. miR-125b regulates calcification of vascular smooth muscle cells. Am. J. Pathol. 179, 1594–1600 (2011).
Liu, J. et al. MicroRNA-32 promotes calcification in vascular smooth muscle cells: implications as a novel marker for coronary artery calcification. PLoS ONE 12, e0174138 (2017).
Wang, S. S., Wang, C. & Chen, H. MicroRNAs are critical in regulating smooth muscle cell mineralization and apoptosis during vascular calcification. J. Cell. Mol. Med. 24, 13564–13572 (2020).
Geekiyanage, H., Rayatpisheh, S., Wohlschlegel, J. A., Brown, R. Jr & Ambros, V. Extracellular microRNAs in human circulation are associated with miRISC complexes that are accessible to anti-AGO2 antibody and can bind target mimic oligonucleotides. Proc. Natl Acad. Sci. USA 117, 24213–24223 (2020).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Vickers, K. C., Palmisano, B. T., Shoucri, B. M., Shamburek, R. D. & Remaley, A. T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 13, 423–433 (2011).
Mittelbrunn, M. et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2, 282 (2011).
Garcia-Martin, R. et al. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 601, 446–451 (2022).
McKenzie, A. J. et al. KRAS-MEK signaling controls Ago2 sorting into exosomes. Cell Rep. 15, 978–987 (2016).
Holland, W. L. et al. Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat. Med. 17, 55–63 (2011).
Turpin, S. M. et al. Obesity-induced CerS6-dependent C16:0 ceramide production promotes weight gain and glucose intolerance. Cell Metab. 20, 678–686 (2014).
Jiang, Y. et al. Peripheral blood miRNAs as a biomarker for chronic cardiovascular diseases. Sci. Rep. 4, 5026 (2014).
Shan, Z. et al. An endocrine genetic signal between blood cells and vascular smooth muscle cells: role of microRNA-223 in smooth muscle function and atherogenesis. J. Am. Coll. Cardiol. 65, 2526–2537 (2015).
Fichtlscherer, S. et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 107, 677–684 (2010).
Brandes, F. et al. Identification of microRNA biomarkers simultaneously expressed in circulating extracellular vesicles and atherosclerotic plaques. Front. Cardiovasc. Med. 11, 1307832 (2024).
Parahuleva, M. S. et al. MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci. Rep. 8, 7823 (2018).
Tabet, F. et al. HDL-transferred microRNA-223 regulates ICAM-1 expression in endothelial cells. Nat. Commun. 5, 3292 (2014).
Vickers, K. C. & Michell, D. L. HDL-small RNA export, transport, and functional delivery in atherosclerosis. Curr. Atheroscler. Rep. 23, 38 (2021).
Mori, M. A., Ludwig, R. G., Garcia-Martin, R., Brandao, B. B. & Kahn, C. R. Extracellular miRNAs: from biomarkers to mediators of physiology and disease. Cell Metab. 30, 656–673 (2019).
Mir, R. et al. Role of selected miRNAs as diagnostic and prognostic biomarkers in cardiovascular diseases, including coronary artery disease, myocardial infarction and atherosclerosis. J. Cardiovasc. Dev. Dis. 8, 22 (2021).
Navickas, R. et al. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc. Res. 111, 322–337 (2016).
Tijsen, A. J., Pinto, Y. M. & Creemers, E. E. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am. J. Physiol. Heart Circ. Physiol. 303, H1085–H1095 (2012).
Barraclough, J. Y., Joan, M., Joglekar, M. V., Hardikar, A. A. & Patel, S. MicroRNAs as prognostic markers in acute coronary syndrome patients — a systematic review. Cells 8, 1572 (2019).
Jakob, P. et al. Profiling and validation of circulating microRNAs for cardiovascular events in patients presenting with ST-segment elevation myocardial infarction. Eur. Heart J. 38, 511–515 (2017).
Saether, J. C. et al. Associations between circulating microRNAs and lipid-rich coronary plaques measured with near-infrared spectroscopy. Sci. Rep. 13, 7580 (2023).
Blanco-Dominguez, R. et al. A novel circulating microRNA for the detection of acute myocarditis. N. Engl. J. Med. 384, 2014–2027 (2021).
Chen, Y., Gao, D. Y. & Huang, L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 81, 128–141 (2015).
Liu, Y. P. & Berkhout, B. miRNA cassettes in viral vectors: problems and solutions. Biochim. Biophys. Acta 1809, 732–745 (2011).
Ye, D. et al. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cell 30, 1645–1654 (2012).
Wang, D. et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat. Cell Biol. 15, 1153–1163 (2013).
Munir, J., Yoon, J. K. & Ryu, S. Therapeutic miRNA-enriched extracellular vesicles: current approaches and future prospects. Cells 9, 2271 (2020).
Pottash, A. E. et al. Combinatorial microRNA loading into extracellular vesicles for increased anti-inflammatory efficacy. Noncoding RNA 8, 71 (2022).
O’Brien, K., Breyne, K., Ughetto, S., Laurent, L. C. & Breakefield, X. O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 21, 585–606 (2020).
Lee, S. W. L. et al. MicroRNA delivery through nanoparticles. J. Control. Release 313, 80–95 (2019).
Brillante, S., Volpe, M. & Indrieri, A. Advances in microRNA therapeutics: from preclinical to clinical studies. Hum. Gene Ther. 35, 628–648 (2024).
Zhou, J. & Rossi, J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 16, 440 (2017).
Hu, S. et al. Mechanisms of hydrogel-based microRNA delivery systems and its application strategies in targeting inflammatory diseases. J. Tissue Eng. 15, 20417314241265897 (2024).
Li, Y. et al. Injectable hydrogel with MSNs/microRNA-21-5p delivery enables both immunomodification and enhanced angiogenesis for myocardial infarction therapy in pigs. Sci. Adv. 7, eabd6740 (2021).
Holjencin, C. & Jakymiw, A. MicroRNAs and their big therapeutic impacts: delivery strategies for cancer intervention. Cells 11, 2332 (2022).
Shang, R., Lee, S., Senavirathne, G. & Lai, E. C. microRNAs in action: biogenesis, function and regulation. Nat. Rev. Genet. 24, 816–833 (2023).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Zhao, L. et al. NONCODEV6: an updated database dedicated to long non-coding RNA annotation in both animals and plants. Nucleic Acids Res. 49, D165–D171 (2021).
Quinn, J. J. et al. Revealing long noncoding RNA architecture and functions using domain-specific chromatin isolation by RNA purification. Nat. Biotechnol. 32, 933–940 (2014).
Carter, A. C. et al. Spen links RNA-mediated endogenous retrovirus silencing and X chromosome inactivation. eLife 9, e54508 (2020).
Sallam, T., Sandhu, J. & Tontonoz, P. Long noncoding RNA discovery in cardiovascular disease: decoding form to function. Circ. Res. 122, 155–166 (2018).
de Goede, O. M. et al. Population-scale tissue transcriptomics maps long non-coding RNAs to complex disease. Cell 184, 2633–2648.e19 (2021).
Liu, S. J. et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 355, aah7111 (2017).
Luo, J., Yang, H. & Song, B. L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 21, 225–245 (2020).
Brown, M. S. & Goldstein, J. L. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell 89, 331–340 (1997).
Horton, J. D., Goldstein, J. L. & Brown, M. S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109, 1125–1131 (2002).
Hong, C. & Tontonoz, P. Liver X receptors in lipid metabolism: opportunities for drug discovery. Nat. Rev. Drug Discov. 13, 433–444 (2014).
Zhang, L. et al. Inhibition of cholesterol biosynthesis through RNF145-dependent ubiquitination of SCAP. eLife 6, e28766 (2017).
Sallam, T. et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature 534, 124–128 (2016).
Zhang, Z. et al. Collaborative interactions of heterogenous ribonucleoproteins contribute to transcriptional regulation of sterol metabolism in mice. Nat. Commun. 11, 984 (2020).
Tontonoz, P. et al. Long noncoding RNA facilitated gene therapy reduces atherosclerosis in a murine model of familial hypercholesterolemia. Circulation 136, 776–778 (2017).
Zhang, Z. et al. A PPARgamma/long noncoding RNA axis regulates adipose thermoneutral remodeling in mice. J. Clin. Invest. 133, e170072 (2023).
Li, C. et al. Regulation of cholesterol homeostasis by a novel long non-coding RNA LASER. Sci. Rep. 9, 7693 (2019).
Wang, Y. et al. Rare variants in long non-coding RNAs are associated with blood lipid levels in the TOPMed whole-genome sequencing study. Am. J. Hum. Genet. 110, 1704–1717 (2023).
Sallam, T. et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 24, 304–312 (2018).
Salisbury, D. A. et al. LncRNAs in inflammation: lessons from a preclinical investigation of Mexis therapy in atherosclerosis. JACC Basic Transl. Sci. 7, 953–955 (2022).
Hennessy, E. J. et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat. Metab. 1, 98–110 (2019).
van Solingen, C. et al. Long noncoding RNA CHROMR regulates antiviral immunity in humans. Proc. Natl Acad. Sci. USA 119, e2210321119 (2022).
Miao, H. et al. A long noncoding RNA distributed in both nucleus and cytoplasm operates in the PYCARD-regulated apoptosis by coordinating the epigenetic and translational regulation. PLoS Genet. 15, e1008144 (2019).
Hu, Y. W. et al. A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J. Lipid Res. 55, 681–697 (2014).
Li, Y., Shen, S., Ding, S. & Wang, L. LincRNA DYN-LRB2-2 upregulates cholesterol efflux by decreasing TLR2 expression in macrophages. J. Cell. Biochem. 119, 1911–1921 (2018).
Cai, C. et al. LncRNA ENST00000602558.1 regulates ABCG1 expression and cholesterol efflux from vascular smooth muscle cells through a p65-dependent pathway. Atherosclerosis 285, 31–39 (2019).
Grabner, G. F., Xie, H., Schweiger, M. & Zechner, R. Lipolysis: cellular mechanisms for lipid mobilization from fat stores. Nat. Metab. 3, 1445–1465 (2021).
Boren, J., Taskinen, M. R., Bjornson, E. & Packard, C. J. Metabolism of triglyceride-rich lipoproteins in health and dyslipidaemia. Nat. Rev. Cardiol. 19, 577–592 (2022).
Ko, C. W., Qu, J., Black, D. D. & Tso, P. Regulation of intestinal lipid metabolism: current concepts and relevance to disease. Nat. Rev. Gastroenterol. Hepatol. 17, 169–183 (2020).
Zhao, X. Y. et al. Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nat. Commun. 9, 2986 (2018).
Zhao, X. Y. et al. The long noncoding RNA Blnc1 orchestrates homeostatic adipose tissue remodeling to preserve metabolic health. Mol. Metab. 14, 60–70 (2018).
Zhao, X. Y., Li, S., Wang, G. X., Yu, Q. & Lin, J. D. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol. Cell 55, 372–382 (2014).
Benhammou, J. N. et al. Novel lipid long intervening noncoding RNA, oligodendrocyte maturation-associated long intergenic noncoding RNA, regulates the liver steatosis gene stearoyl-coenzyme A desaturase as an enhancer RNA. Hepatol. Commun. 3, 1356–1372 (2019).
Jiang, Y. et al. Loss of Hilnc prevents diet-induced hepatic steatosis through binding of IGF2BP2. Nat. Metab. 3, 1569–1584 (2021).
Wang, J. et al. The novel long noncoding RNA Lnc19959.2 modulates triglyceride metabolism-associated genes through the interaction with Purb and hnRNPA2B1. Mol. Metab. 37, 100996 (2020).
Huang, P. et al. LncRNA MEG3 functions as a ceRNA in regulating hepatic lipogenesis by competitively binding to miR-21 with LRP6. Metabolism 94, 1–8 (2019).
Schoeler, M. & Caesar, R. Dietary lipids, gut microbiota and lipid metabolism. Rev. Endocr. Metab. Disord. 20, 461–472 (2019).
Wang, Y. et al. The gut microbiota reprograms intestinal lipid metabolism through long noncoding RNA Snhg9. Science 381, 851–857 (2023).
Guo, Y. F. et al. lncRNA Hnscr regulates lipid metabolism by mediating adipocyte lipolysis. Endocrinology 164, bqad147 (2023).
Liu, B. et al. LncRNA Nron deficiency protects mice from diet-induced adiposity and hepatic steatosis. Metabolism 148, 155609 (2023).
Han, L. et al. Lipid droplet-associated lncRNA LIPTER preserves cardiac lipid metabolism. Nat. Cell Biol. 25, 1033–1046 (2023).
Zhang, X. et al. Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci. Transl. Med. 10, eaar5987 (2018).
Chen, J. et al. Deficiency of lncRNA MERRICAL abrogates macrophage chemotaxis and diabetes-associated atherosclerosis. Cell Rep. 43, 113815 (2024).
Cynn, E. et al. Human macrophage long intergenic noncoding RNA, SIMALR, suppresses inflammatory macrophage apoptosis via NTN1 (Netrin-1). Arterioscler. Thromb. Vasc. Biol. 43, 286–299 (2023).
Xiang, D. et al. Leukocyte-specific Morrbid promotes leukocyte differentiation and atherogenesis. Research 6, 0187 (2023).
Tanwar, V. S. et al. Palmitic acid-induced long noncoding RNA PARAIL regulates inflammation via interaction with RNA-binding protein ELAVL1 in monocytes and macrophages. Arterioscler. Thromb. Vasc. Biol. 43, 1157–1175 (2023).
Kotzin, J. J. et al. The long non-coding RNA Morrbid regulates Bim and short-lived myeloid cell lifespan. Nature 537, 239–243 (2016).
Karasawa, T. et al. Saturated fatty acids undergo intracellular crystallization and activate the NLRP3 inflammasome in macrophages. Arterioscler. Thromb. Vasc. Biol. 38, 744–756 (2018).
Ma, Y., Jiang, C. F., Li, P. & Cao, H. In vivo functional analysis of nonconserved human lncRNAs using a humanized mouse model. Methods Mol. Biol. 2254, 339–347 (2021).
Zilbauer, M. et al. A roadmap for the human gut cell atlas. Nat. Rev. Gastroenterol. Hepatol. 20, 597–614 (2023).
Zhao, Z. et al. Organoids. Nat. Rev. Methods Primers 2, 94 (2022).
Qian, X., Zhao, J., Yeung, P. Y., Zhang, Q. C. & Kwok, C. K. Revealing lncRNA structures and interactions by sequencing-based approaches. Trends Biochem. Sci. 44, 33–52 (2019).
Economides, A. N. et al. Conditionals by inversion provide a universal method for the generation of conditional alleles. Proc. Natl Acad. Sci. USA 110, E3179–E3188 (2013).
Lyu, Q. R., Zhang, S., Zhang, Z. & Tang, Z. Functional knockout of long non-coding RNAs with genome editing. Front. Genet. 14, 1242129 (2023).
Zhang, S. et al. BESST: a novel LncRNA knockout strategy with less genome perturbance. Nucleic Acids Res. 51, e49 (2023).
Putnam, A., Thomas, L. & Seydoux, G. RNA granules: functional compartments or incidental condensates? Genes Dev. 37, 354–376 (2023).
Han, T. W. et al. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell 149, 768–779 (2012).
Tian, S., Curnutte, H. A. & Trcek, T. RNA granules: a view from the RNA perspective. Molecules 25, 3130 (2020).
Onoguchi-Mizutani, R. & Akimitsu, N. Long noncoding RNA and phase separation in cellular stress response. J. Biochem. 171, 269–276 (2022).
Kan, R. L., Chen, J. & Sallam, T. Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation. Trends Genet. 38, 182–193 (2022).
Jiang, X. et al. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 6, 74 (2021).
Liu, J. et al. N6-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription. Science 367, 580–586 (2020).
Wei, J. et al. FTO mediates LINE1 m(6)A demethylation and chromatin regulation in mESCs and mouse development. Science 376, 968–973 (2022).
Uzonyi, A. et al. Exclusion of m6A from splice-site proximal regions by the exon junction complex dictates m6A topologies and mRNA stability. Mol. Cell 83, 237–251 e237 (2023).
Vicens, Q. & Kieft, J. S. Thoughts on how to think (and talk) about RNA structure. Proc. Natl Acad. Sci. USA 119, e2112677119 (2022).
Kim, Y. & Lee, M. Deep learning approaches for lncRNA-mediated mechanisms: a comprehensive review of recent developments. Int. J. Mol. Sci. 24, 10299 (2023).
Irani, S., Iqbal, J., Antoni, W. J., Ijaz, L. & Hussain, M. M. microRNA-30c reduces plasma cholesterol in homozygous familial hypercholesterolemic and type 2 diabetic mouse models. J. Lipid Res. 59, 144–154 (2018).
Wang, L. et al. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol. Cell. Biol. 33, 1956–1964 (2013).
Hu, Z., Shen, W. J., Kraemer, F. B. & Azhar, S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol. Cell. Biol. 32, 5035–5045 (2012).
Kang, M. H. et al. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler. Thromb. Vasc. Biol. 33, 2724–2732 (2013).
Chen, C., Matye, D., Wang, Y. & Li, T. Liver-specific microRNA-185 knockout promotes cholesterol dysregulation in mice. Liver Res. 5, 232–238 (2021).
Yang, M. et al. Identification of miR-185 as a regulator of de novo cholesterol biosynthesis and low density lipoprotein uptake. J. Lipid Res. 55, 226–238 (2014).
Jiang, Y., Yin, H. & Zheng, X. L. MicroRNA-1 inhibits myocardin-induced contractility of human vascular smooth muscle cells. J. Cell. Physiol. 225, 506–511 (2010).
Sun, S. G. et al. miR-146a and Krüppel-like factor 4 form a feedback loop to participate in vascular smooth muscle cell proliferation. EMBO Rep. 12, 56–62 (2011).
Halley, P. et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 6, 222–230 (2014).
Ruan, X. et al. Identification of human long noncoding RNAs associated with nonalcoholic fatty liver disease and metabolic homeostasis. J. Clin. Invest. 131, e136336 (2021).
Holdt, L. M. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7, 12429 (2016).
Vacante, F. et al. CARMN loss regulates smooth muscle cells and accelerates atherosclerosis in mice. Circ. Res. 128, 1258–1275 (2021).
Zhang, W. et al. INKILN is a novel long noncoding RNA promoting vascular smooth muscle inflammation via scaffolding MKL1 and USP10. Circulation 148, 47–67 (2023).
Li, S. et al. LncRNA PSMB8-AS1 instigates vascular inflammation to aggravate atherosclerosis. Circ. Res. 134, 60–80 (2024).
Zhang, R. et al. LncRNA RP4-639F20.1 interacts with THRAP3 to attenuate atherosclerosis by regulating c-FOS in vascular smooth muscle cells proliferation and migration. Atherosclerosis 379, 117183 (2023).
Agbu, P. et al. MicroRNA-mediated regulation of glucose and lipid metabolism. Nat. Rev. Mol. Cell Biol. 22, 425–438 (2021).
Libby, P. et al. Atherosclerosis. Nat. Rev. Dis. Primers 5, 56 (2019).
Chen, W. Macrophage-targeted nanomedicine for the diagnosis and treatment of atherosclerosis. Nat. Rev. Cardiol. 19, 228–249 (2022).
Acknowledgements
The authors are supported by grants from the American Heart Association (23SCEFIA1153739 to C.v.S.) and the National Institutes of Health (R01HL172335, R01HL172365 and P01HL131481 to K.J.M.; and R01 DK957554, R01 HL139549 and R01HL149766 to T.S.).
Author information
Authors and Affiliations
Contributions
The authors contributed substantially to all aspects of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors hold patents on the use of inhibitors targeting miR-33 (K.J.M.), CHROMR (K.J.M. and C.v.S.) and RALY (T.S.).
Peer review
Peer review information
Nature Reviews Cardiology thanks Alberto Davalos, M. Mahmood Hussain and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Antisense oligonucleotides
-
(ASOs). Short, synthetic RNA strands designed to bind a specific RNA sequence and alter its stability.
- Aptamer
-
Short, structured nucleic acid ligands that fold into defined shapes to bind specific proteins or small molecules with high affinity.
- Cholesterol efflux
-
The cellular export of cholesterol to extracellular acceptors, primarily apolipoprotein A-1-containing HDL, via transporters such as phospholipid-transporting ATPase ABCA1.
- Extracellular miRNAs
-
MicroRNAs (miRNAs) released from cells within vesicles or bound to proteins, which can be taken up by recipient cells to modulate gene expression.
- HDL
-
Apolipoprotein A1-containing lipoprotein particles that mediate reverse cholesterol transport from tissues back to the liver.
- Hepatosteatosis
-
Accumulation of fat droplets within hepatocytes, commonly referred to as fatty liver.
- LDL
-
Cholesterol-rich particles that deliver cholesterol from the liver to peripheral tissues.
- Macrophage foam cells
-
Lipid-laden macrophages in the arterial intima that arise from macrophage uptake of modified lipoproteins and that contribute to atherosclerotic plaque formation.
- Reverse cholesterol transport
-
Multistep process by which cholesterol is removed from peripheral tissues and delivered back to the liver for excretion.
- Vascular smooth muscle cells
-
(VSMCs). Phenotypic switching of vascular smooth muscle cells into alternative cell types (such as macrophage-like or osteogenic) that contribute to atherosclerosis.
- VLDL
-
Triglyceride-rich particles secreted by the liver that transport endogenous lipids to peripheral tissues.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sallam, T., van Solingen, C. & Moore, K.J. Non-coding RNAs in lipid metabolism and their roles in atherosclerosis. Nat Rev Cardiol (2026). https://doi.org/10.1038/s41569-025-01229-9
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41569-025-01229-9