Abstract
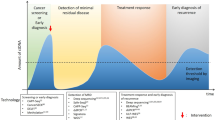
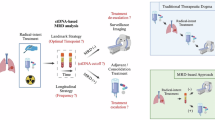
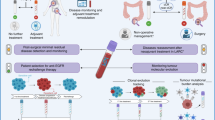
Metastasis is the leading cause of cancer-related death in patients with solid tumours. Current imaging technologies are not sufficiently sensitive to detect minimal residual disease (MRD; also known as measurable or molecular residual disease) after initial surgery or chemotherapy, pointing to the need for more sensitive tests to detect remaining traces of cancer in the body. Liquid biopsy, or the analysis of tumour-derived or tumour-induced cells or cellular products in the blood or other body fluids, has opened a new diagnostic avenue to detect and monitor MRD. Liquid biopsy is already used in clinical decision making for patients with haematological malignancies. Here, we review current knowledge on the use of circulating tumour DNA (ctDNA) to detect and monitor MRD in patients with solid tumours. We also discuss how ctDNA-guided MRD detection and characterization could herald a new era of novel ‘post-adjuvant therapies’ with the potential to eliminate MRD and cure patients before terminal metastatic disease is evident on imaging.
Key points
-
Sensitive blood assays enable the detection of circulating tumour DNA (ctDNA) at very low concentrations (down to <10 ppm).
-
The detection of very low concentrations of ctDNA is required to detect minimal residual disease (MRD; also known as measurable or molecular residual disease) in patients with solid tumours.
-
MRD detection can be correlated with clinical outcomes such as progression-free survival and overall survival.
-
ctDNA-based MRD detection can precede imaging-based detection of relapse by several months depending on the tumour type and stage.
-
The transition of ctDNA-detectable MRD to overt metastasis can depend on the residual tumour burden and the host response (for example, antitumour immunity).
-
Interventional clinical studies in patients with solid tumours and MRD positivity are now required to assess which treatment(s) can eliminate MRD in these patients.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
van der Velden, V. H. J. et al. Analysis of measurable residual disease by IG/TR gene rearrangements: quality assurance and updated EuroMRD guidelines. Leukemia 38, 1315–1322 (2024).
Walter, R. B. & Gale, R. P. Measurable residual disease in haematological and solid cancers. Leukemia 38, 1647–1648 (2024).
Pantel, K. & Alix-Panabieres, C. Liquid biopsy and minimal residual disease – latest advances and implications for cure. Nat. Rev. Clin. Oncol. 16, 409–424 (2019).
Pich, O., Reyes-Salazar, I., Gonzalez-Perez, A. & Lopez-Bigas, N. Discovering the drivers of clonal hematopoiesis. Nat. Commun. 13, 4267 (2022).
Kato, S., Lippman, S. M., Flaherty, K. T. & Kurzrock, R. The conundrum of genetic “drivers” in benign conditions. J. Natl Cancer Inst. 108, https://doi.org/10.1093/jnci/djw036 (2016).
Nishimura, T. et al. Evolutionary histories of breast cancer and related clones. Nature 620, 607–614 (2023).
Reed, S. C., Croessmann, S. & Park, B. H. CHIP happens: clonal hematopoiesis of indeterminate potential and its relationship to solid tumors. Clin. Cancer Res. 29, 1403–1411 (2023).
Henriksen, T. V. et al. Unraveling the potential clinical utility of circulating tumor DNA detection in colorectal cancer-evaluation in a nationwide Danish cohort. Ann. Oncol. 35, 229–239 (2024).
Rehman, A. U. et al. Liquid biopsies to occult brain metastasis. Mol. Cancer 21, 113 (2022).
Bettegowda, C. et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 6, 224ra224 (2014).
Merker, J. D. et al. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 36, 1631–1641 (2018).
Diehl, F. et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 14, 985–990 (2008).
Rostami, A. et al. Senescence, necrosis, and apoptosis govern circulating cell-free DNA release kinetics. Cell Rep. 31, 107830 (2020).
Mattox, A. K. et al. The origin of highly elevated cell-free DNA in healthy individuals and patients with pancreatic, colorectal, lung, or ovarian cancer. Cancer Discov. 13, 2166–2179 (2023).
Martin-Alonso, C. et al. Priming agents transiently reduce the clearance of cell-free DNA to improve liquid biopsies. Science 383, eadf2341 (2024).
Cinar, M. et al. Transposon DNA sequences facilitate the tissue-specific gene transfer of circulating tumor DNA between human cells. Nucleic Acids Res. 52, 7539–7555 (2024).
Malkin, E. Z. et al. Cell-free DNA topology depends on its subcellular and cellular origins in cancer. JCI Insight 7, e159590 (2022).
Parikh, A. R. et al. Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin. Cancer Res. 27, 5586–5594 (2021).
Schrag, D. et al. Blood-based tests for multicancer early detection (PATHFINDER): a prospective cohort study. Lancet 402, 1251–1260 (2023).
Nakamura, Y. et al. Longitudinal clinical performance of a novel tumor-naive minimal residual disease assay in patients with resected stage II and III colorectal cancer: a subset analysis from the GALAXY study in CIRCULATE-Japan. J. Clin. Oncol. 42, 3618–3618 (2024).
Heitzer, E., Haque, I. S., Roberts, C. E. S. & Speicher, M. R. Current and future perspectives of liquid biopsies in genomics-driven oncology. Nat. Rev. Genet. 20, 71–88 (2019).
Moser, T., Kühberger, S., Lazzeri, I., Vlachos, G. & Heitzer, E. Bridging biological cfDNA features and machine learning approaches. Trends Genet. 39, 285–307 (2023).
Gydush, G. et al. Massively parallel enrichment of low-frequency alleles enables duplex sequencing at low depth. Nat. Biomed. Eng. 6, 257–266 (2022).
Parsons, H. A. et al. Circulating tumor DNA association with residual cancer burden after neoadjuvant chemotherapy in triple-negative breast cancer in TBCRC 030. Ann. Oncol. 34, 899–906 (2023).
Zheng, J., Qin, C., Wang, Q., Tian, D. & Chen, Z. Circulating tumour DNA-based molecular residual disease detection in resectable cancers: a systematic review and meta-analysis. EBioMedicine 103, 105109 (2024).
Henriksen, T. V. et al. Circulating tumor DNA in stage III colorectal cancer, beyond minimal residual disease detection, toward assessment of adjuvant therapy efficacy and clinical behavior of recurrences. Clin. Cancer Res. 28, 507–517 (2022).
Zhong, R. et al. Accuracy of minimal residual disease detection by circulating tumor DNA profiling in lung cancer: a meta-analysis. BMC Med. 21, 180 (2023).
Taieb, J. et al. Prognostic value and relation with adjuvant treatment duration of ctDNA in stage III colon cancer: a post hoc analysis of the PRODIGE-GERCOR IDEA-France trial. Clin. Cancer Res. 27, 5638–5646 (2021).
Mo, S. et al. Early detection of molecular residual disease and risk stratification for stage I to III colorectal cancer via circulating tumor DNA methylation. JAMA Oncol. 9, 770–778 (2023).
Tie, J. et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N. Engl. J. Med. 386, 2261–2272 (2022).
Tie, J., Lo, S. N. & Gibbs, P. Circulating tumor DNA guiding adjuvant therapy in colon cancer. reply. N. Engl. J. Med. 387, 760 (2022).
Tie, J. et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer: overall survival and updated 5-year results from the randomized DYNAMIC trial. J. Clin. Oncol. 42, 16 (2024).
Lonardi, S. et al. The PEGASUS trial: post-surgical liquid biopsy-guided treatment of stage III and high-risk stage II colon cancer patients [abstract LBA28]. Ann. Oncol. 34 (Suppl. 2), 1268–1269 (2023).
Kotani, D. et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat. Med. 29, 127–134 (2023).
Köhne, C. H. et al. FOLFIRI plus cetuximab in patients with liver-limited or non-liver-limited RAS wild-type metastatic colorectal cancer: a retrospective subgroup analysis of the CRYSTAL study. Eur. J. Surg. Oncol. 42, 1540–1547 (2016).
Newhook, T. E. et al. Prospective study of perioperative circulating tumor DNA dynamics in patients undergoing hepatectomy for colorectal liver metastases. Ann. Surg. 277, 813–820 (2023).
Jiang, H. et al. Postoperative circulating tumor DNA testing based on tumor naïve strategy after liver metastasis surgery in colorectal cancer patients. Front. Oncol. 13, 1153685 (2023).
Nors, J. et al. IMPROVE-IT2: implementing noninvasive circulating tumor DNA analysis to optimize the operative and postoperative treatment for patients with colorectal cancer–intervention trial 2. Study protocol. Acta Oncol. 59, 336–341 (2020).
Symonds, E. L. et al. Assessment of tumor burden and response to therapy in patients with colorectal cancer using a quantitative ctDNA test for methylated BCAT1/IKZF1. Mol. Oncol. 16, 2031–2041 (2022).
Maddalena, G. et al. INTERCEPT program of circulating tumor DNA (ctDNA) testing for minimal residual disease (MRD) in colorectal cancer (CRC): results from a prospective clinical cohort [abstract]. J. Clin. Oncol. 42 (Suppl. 3), 27 (2024).
Eluri, M. et al. Short lead time and high rates of concomitant radiographic recurrences for ctDNA-based minimal residual disease assays colorectal cancer (CRC) during surveillance: results from the MD Anderson INTERCEPT program [abstract 587P]. Ann. Oncol. 34 (Suppl. 2), 427 (2023).
Abbosh, C. et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 616, 553–562 (2023).
Gale, D. et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 33, 500–510 (2022).
Zhang, J. T. et al. Longitudinal undetectable molecular residual disease defines potentially cured population in localized non-small cell lung cancer. Cancer Discov. 12, 1690–1701 (2022).
Chen, K. et al. Individualized tumor-informed circulating tumor DNA analysis for postoperative monitoring of non-small cell lung cancer. Cancer Cell 41, 1749–1762.e6 (2023).
Felip, E. et al. IMpower010: ctDNA status in patients (pts) with resected NSCLC who received adjuvant chemotherapy (chemo) followed by atezolizumab (atezo) or best supportive care (BSC) [abstract 1O]. Immunooncol. Technol. 16 (Suppl. 1), 1–2 (2022).
Reck, M. et al. Associations of ctDNA clearance and pathological response with neoadjuvant treatment in patients with resectable NSCLC from the phase III AEGEAN trial [abstract LBA59]. Ann. Oncol. 34 (Suppl. 2), S1300 (2023).
Provencio, M. et al. Overall survival and biomarker analysis of neoadjuvant nivolumab plus chemotherapy in operable stage IIIA non-small-cell lung cancer (NADIM phase II trial). J. Clin. Oncol. 40, 2924–2933 (2022).
Pantel, K. et al. GUIDE.MRD: A Consortium guiding multi-modal therapies against minimal residual disease (MRD) by liquid biopsy to assess implementation of circulating tumor DNA (ctDNA) in clinical practice to improve patient outcomes [abstract 237TiP]. Ann. Oncol. 34 (Suppl. 2), 276–S277 (2023).
Prat, A. et al. Circulating tumor DNA reveals complex biological features with clinical relevance in metastatic breast cancer. Nat. Commun. 14, 1157 (2023).
Turner, N. C. et al. Results of the c-TRAK TN trial: a clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann. Oncol. 34, 200–211 (2023).
Garcia-Murillas, I. et al. Ultra-sensitive ctDNA mutation tracking to identify molecular residual disease and predict relapse in patients with early breast cancer. J. Clin. Oncol. 42 (Suppl. 16), 1010 (2024).
Bidard, F. C. et al. Elacestrant (oral selective estrogen receptor degrader) versus standard endocrine therapy for estrogen receptor-positive, human epidermal growth factor receptor 2-negative advanced breast cancer: results from the randomized phase III EMERALD trial. J. Clin. Oncol. 40, 3246–3256 (2022).
Pan, H. et al. 20-year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N. Engl. J. Med. 377, 1836–1846 (2017).
Hennigan, S. T. et al. Low abundance of circulating tumor DNA in localized prostate cancer. JCO Precis. Oncol. 3, https://doi.org/10.1200/po.19.00176 (2019).
Trujillo, B., Wu, A., Wetterskog, D. & Attard, G. Blood-based liquid biopsies for prostate cancer: clinical opportunities and challenges. Br. J. Cancer 127, 1394–1402 (2022).
Kasi, P. M. et al. Circulating tumor DNA enables sensitive detection of actionable gene fusions and rearrangements across cancer types. Clin. Cancer Res. 30, 836–848 (2024).
Fei, X. et al. Early plasma circulating tumor DNA as a potential biomarker of disease recurrence in non-metastatic prostate cancer. Cancer Res. Treat. 55, 969–977 (2023).
Powles, T. et al. ctDNA guiding adjuvant immunotherapy in urothelial carcinoma. Nature 595, 432–437 (2021).
Powles, T. et al. Updated overall survival by circulating tumor DNA status from the phase 3 IMvigor010 trial: adjuvant atezolizumab versus observation in muscle-invasive urothelial carcinoma. Eur. Urol. 85, 114–122 (2024).
Territo, A. et al. DNA methylation urine biomarkers test in the diagnosis of upper tract urothelial carcinoma: results from a single-center prospective clinical trial. J. Urol. 208, 570–579 (2022).
Pierconti, F. et al. Upper urothelial tract high-grade carcinoma: comparison of urine cytology and DNA methylation analysis in urinary samples. Hum. Pathol. 118, 42–48 (2021).
Yuan, S. Q. et al. Residual circulating tumor DNA after adjuvant chemotherapy effectively predicts recurrence of stage II-III gastric cancer. Cancer Commun. 43, 1312–1325 (2023).
Guo, D.-Z. et al. Utilization of tumor-informed circulating tumor DNA in detecting minimal residual disease and guiding adjuvant therapy in liver cancer [abstract]. J. Clin. Oncol. 42 (Suppl. 16), 4125 (2024).
Lee, B. et al. The potential role of serial circulating tumor DNA (ctDNA) testing after upfront surgery to guide adjuvant chemotherapy for early stage pancreatic cancer: the AGITG DYNAMIC-Pancreas trial [abstract]. J. Clin. Oncol. 42 (Suppl. 16), 107 (2024).
Han, K. et al. Clinical validation of human papilloma virus circulating tumor DNA for early detection of residual disease after chemoradiation in cervical cancer. J. Clin. Oncol. 42, 431–440 (2024).
Herbst, J. et al. Detection of multiple HPV types in liquid biopsies of cervical neoplasia. Clin. Chem. 70, 285–296 (2024).
Lee, N. Y., Morris, L. G. T. & Diehn, M. Assessing the evidence for circulating tumor HPV DNA in patients with oropharyngeal cancer. JAMA Oncol. 10, 1021–1022 (2024).
Sanz-Garcia, E. et al. Multimodal detection of molecular residual disease in high-risk locally advanced squamous cell carcinoma of the head and neck. Cell Death Differ. 31, 460–468 (2024).
Zhu, J. W. et al. Evaluating the utility of ctDNA in detecting residual cancer and predicting recurrence in patients with serous ovarian cancer. Int. J. Mol. Sci. 24, 14388 (2023).
Heo, J. et al. Serial circulating tumor DNA analysis with a tumor-naïve next-generation sequencing panel detects minimal residual disease and predicts outcome in ovarian cancer. Cancer Res. 84, 468–478 (2024).
Alix-Panabieres, C. & Pantel, K. Liquid biopsy: from discovery to clinical application. Cancer Discov. 11, 858–873 (2021).
Woo, H. J. et al. Continuous centrifugal microfluidics (CCM) isolates heterogeneous circulating tumor cells via full automation. Theranostics 12, 3676–3689 (2022).
Lucien, F. et al. MIBlood-EV: minimal information to enhance the quality and reproducibility of blood extracellular vesicle research. J. Extracell. Vesicles 12, e12385 (2023).
Linville, L. M. et al. Utility of circulating tumor DNA (ctDNA) to inform treatment of patients with metastatic breast cancer [abstract]. J. Clin. Oncol. 42 (Suppl. 16), 1042 (2024).
Stergiopoulou, D. et al. Comprehensive liquid biopsy analysis as a tool for the early detection of minimal residual disease in breast cancer. Sci. Rep. 13, 1258 (2023).
Zhao, L. et al. Integrated analysis of circulating tumour cells and circulating tumour DNA to detect minimal residual disease in hepatocellular carcinoma. Clin. Transl. Med. 12, e793 (2022).
Eslami, S. Z. et al. Circulating tumour cells and PD-L1-positive small extracellular vesicles: the liquid biopsy combination for prognostic information in patients with metastatic non-small cell lung cancer. Br. J. Cancer 130, 63–72 (2024).
Mergel, F. et al. SURVIVE study – a multicenter, randomized, controlled phase 3 superiority trial, evaluating liquid biopsy guided intensified follow-up surveillance in women with intermediate-to high-risk early breast cancer [abstract]. Cancer Res. 84 (Suppl. 9), PO1-20-05 (2024).
Pfister, K. et al. The SURVIVE study: liquid biopsy guided surveillance after intermediate- to high-risk early breast cancer [abstract]. J. Clin. Oncol. 42 (Suppl. 16), TPS620 (2024).
Lipsyc-Sharf, M. et al. Circulating tumor DNA and late recurrence in high-risk hormone receptor-positive, human epidermal growth factor receptor 2-negative breast cancer. J. Clin. Oncol. 40, 2408–2419 (2022).
Rajan, N., Khanal, T. & Ringel, M. D. Progression and dormancy in metastatic thyroid cancer: concepts and clinical implications. Endocrine 70, 24–35 (2020).
Singvogel, K. & Schittek, B. Dormancy of cutaneous melanoma. Cancer Cell Int. 24, 88 (2024).
Pinto-Coelho, L. How artificial intelligence is shaping medical imaging technology: a survey of innovations and applications. Bioengineering 10, https://doi.org/10.3390/bioengineering10121435 (2023).
Kandel, S. et al. Demonstration of an AI-driven workflow for autonomous high-resolution scanning microscopy. Nat. Commun. 14, 5501 (2023).
Khalifa, M. & Albadawy, M. AI in diagnostic imaging: revolutionising accuracy and efficiency. Comput. Methods Prog. Biomed. Update 5, 100146 (2024).
Song, A. H. et al. Analysis of 3D pathology samples using weakly supervised AI. Cell 187, 2502–2520.e17 (2024).
Xu, H. et al. A whole-slide foundation model for digital pathology from real-world data. Nature 630, 181–188 (2024).
You, Y. et al. Systematic comparison of sequencing-based spatial transcriptomic methods. Nat. Methods 21, 1743–1754 (2024).
Benjamin, K. et al. Multiscale topology classifies cells in subcellular spatial transcriptomics. Nature 630, 943–949 (2024).
Bando, H. et al. Effects of metastatic sites on circulating tumor DNA in patients with metastatic colorectal cancer. JCO Precis. Oncol. 6, e2100535 (2022).
Dong, S. et al. Circulating tumor DNA-guided de-escalation targeted therapy for advanced non-small cell lung cancer: a nonrandomized clinical trial. JAMA Oncol. 10, 932–940 (2024).
Stadler, J. C. et al. Prognostic value of von Willebrand factor levels in patients with metastatic melanoma treated by immune checkpoint inhibitors. J. Immunother. Cancer 11, e006456 (2023).
Loyfer, N. et al. A DNA methylation atlas of normal human cell types. Nature 613, 355–364 (2023).
Pantel, K. & Hayes, D. F. Disseminated breast tumour cells: biological and clinical meaning. Nat. Rev. Clin. Oncol. 15, 129–131 (2018).
Wan, J. C. M. et al. Liquid biopsies for residual disease and recurrence. Med 2, 1292–1313 (2021).
Braun, S. et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N. Engl. J. Med. 353, 793–802 (2005).
Tivey, A., Church, M., Rothwell, D., Dive, C. & Cook, N. Circulating tumour DNA – looking beyond the blood. Nat. Rev. Clin. Oncol. 19, 600–612 (2022).
Chauhan, P. S. et al. Urine cell-free DNA multi-omics to detect MRD and predict survival in bladder cancer patients. NPJ Precis. Oncol. 7, 6 (2023).
Darlix, A. et al. Detection of circulating tumor cells in cerebrospinal fluid of patients with suspected breast cancer leptomeningeal metastases: a prospective study. Clin. Chem. 68, 1311–1322 (2022).
Escudero, L. et al. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat. Commun. 11, 5376 (2020).
Chai, R. et al. Sequencing of cerebrospinal fluid cell-free DNA facilitated early differential diagnosis of intramedullary spinal cord tumors. NPJ Precis. Oncol. 8, 43 (2024).
Connors, D. et al. International Liquid Biopsy Standardization Alliance white paper. Crit. Rev. Oncol. Hematol. 156, 103112 (2020).
Hayes, D. F. Biomarker validation and testing. Mol. Oncol. 9, 960–966 (2015).
Acknowledgements
K.P. received funding from the Deutsche Forschungsgemeinschaft Priority Program SPP 2084 microBONE (DFG), Deutsche Krebshilfe project CUPIDO and the European Research Council (ERC) Advanced Investigator Grant INJURMET (no. 834974). C.A.-P. received funding from La Fondation ARC pour la Recherche sur le Cancer with the project “PANLIPSY” on liquid biopsy and pancreatic cancer, the Institut National du Cancer and from les Fonds de Dotation AFER pour la Recherche Médicale.
Author information
Authors and Affiliations
Contributions
The authors contributed equally to all aspects of the article preparation.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks A. A. Chaudhuri, Y. Nakamura, who co-reviewed with T. Hashimoto, C. Rolfo and A. Sartore-Bianchi for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pantel, K., Alix-Panabières, C. Minimal residual disease as a target for liquid biopsy in patients with solid tumours. Nat Rev Clin Oncol 22, 65–77 (2025). https://doi.org/10.1038/s41571-024-00967-y
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41571-024-00967-y