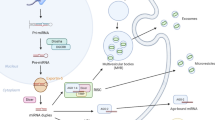
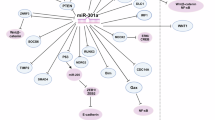
Abstract
Over the past three decades, knowledge of microRNA (miRNA) biology has advanced from the initial discovery of their regulatory functions to the finding of abnormal activity in leukaemias, and then to a comprehensive understanding of the roles of miRNAs in both normal physiology and most diseases, with cancer being extensively studied. miRNA dysregulation contributes to tumorigenesis, with certain miRNAs acting as either tumour suppressors or oncogenic factors in a context-dependent manner. A subset of miRNAs have shown promise as tumour biomarkers and therapeutic targets in preclinical studies, with several miRNA-based diagnostic tools and treatments progressing to clinical trials. Artificial intelligence (AI) and machine learning techniques began to be introduced into cancer research and oncology a decade ago and are now on the verge of revolutionizing biomarker identification and clinical trials. In this Review, we highlight important roles of miRNAs in cancer biology and their potential as diagnostic tools and therapeutic targets. In particular, we discuss emerging challenges and opportunities presented by AI-driven data analysis and combinatorial strategies, and how advances in these areas have addressed previous doubts on the clinical translation of miRNA-based biomarkers and therapeutics.
Key points
-
Despite extensive studies, microRNAs (miRNAs) are not used clinically as biomarkers, owing to insufficient diagnostic specificity and sensitivity.
-
Contemporary studies of miRNA biomarkers aim to improve diagnostic tools by using miRNA profiles, considering miRNA transport mechanisms, integrating miRNA and other omics data, and applying machine learning (ML) and other artificial intelligence (AI) methods to integrate the data.
-
AI and ML enable complex data analysis to identify multi-miRNA signatures for cancer subtyping, early detection and pan-cancer diagnostics using models such as random forests, deep learning and foundation models.
-
AI and ML tools are also advancing the development of miRNA-based therapeutics, although challenges relating to data quality and algorithm transparency highlight the need for standardized, collaborative and interpretable frameworks for reliable translational research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Ambros, V. The functions of animal microRNAs. Nature 431, 350–355 (2004).
Drula, R. & Calin, G. A. MicroRNAs: where brilliance, perseverance, and ambition converged. J. Clin. Invest. 135, e189625 (2024).
Stefani, G. & Slack, F. J. Small non-coding RNAs in animal development. Nat. Rev. Mol. Cell Biol. 9, 219–230 (2008).
Dragomir, M. P., Knutsen, E. & Calin, G. A. Classical and noncanonical functions of miRNAs in cancers. Trends Genet. 38, 379–394 (2022).
Calin, G. A. et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA 99, 15524–15529 (2002).
Martino, M. T. D., Tagliaferri, P. & Tassone, P. MicroRNA in cancer therapy: breakthroughs and challenges in early clinical applications. J. Exp. Clin. Cancer Res. 44, 126 (2025).
Winkle, M., El-Daly, S. M., Fabbri, M. & Calin, G. A. Noncoding RNA therapeutics — challenges and potential solutions. Nat. Rev. Drug Discov. 20, 629–651 (2021).
Slack, F. J. & Chinnaiyan, A. M. The role of non-coding RNAs in oncology. Cell 179, 1033–1055 (2019).
Tentori, A. M. et al. Quantitative and multiplex microRNA assays from unprocessed cells in isolated nanoliter well arrays. Lab. Chip 18, 2410–2424 (2018).
Alvarez-Torres, M.dM., Fu, X. & Rabadan, R. Illuminating the noncoding genome in cancer using artificial intelligence. Cancer Res. 85, 2368–2375 (2025).
Shenoy, A. & Blelloch, R. H. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat. Rev. Mol. Cell Biol. 15, 565–576 (2014).
Davis, B. N. & Hata, A. Regulation of microRNA biogenesis: a miRiad of mechanisms. Cell Commun. Signal. 7, 18 (2009).
Blow, M. J. et al. RNA editing of human microRNAs. Genome Biol. 7, R27 (2006).
Hwang, H. W., Wentzel, E. A. & Mendell, J. T. Cell–cell contact globally activates microRNA biogenesis. Proc. Natl Acad. Sci. USA 106, 7016–7021 (2009).
Kwanhian, W. et al. MicroRNA-142 is mutated in about 20% of diffuse large B-cell lymphoma. Cancer Med. 1, 141–155 (2012).
Bracken, C. P., Scott, H. S. & Goodall, G. J. A network-biology perspective of microRNA function and dysfunction in cancer. Nat. Rev. Genet. 17, 719–732 (2016).
Broseghini, E., Dika, E., Londin, E. & Ferracin, M. MicroRNA isoforms contribution to melanoma pathogenesis. Noncoding RNA 7, 63 (2021).
Guo, H., Zhang, N., Huang, T. & Shen, N. MicroRNA-200c in cancer generation, invasion, and metastasis. Int. J. Mol. Sci. 26, 710 (2025).
Di Leva, G. & Croce, C. M. Roles of small RNAs in tumor formation. Trends Mol. Med. 16, 257–267 (2010).
Chirshev, E., Oberg, K. C., Ioffe, Y. J. & Unternaehrer, J. J. Let-7 as biomarker, prognostic indicator, and therapy for precision medicine in cancer. Clin. Transl. Med. 8, 24 (2019).
Baer, C. et al. Suppression of microRNA activity amplifies IFN-γ-induced macrophage activation and promotes anti-tumour immunity. Nat. Cell Biol. 18, 790–802 (2016).
Hwang, H. W., Wentzel, E. A. & Mendell, J. T. A hexanucleotide element directs microRNA nuclear import. Science 315, 97–100 (2007).
Fabbri, M. et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl Acad. Sci. USA 109, E2110–E2116 (2012).
Setrerrahmane, S. et al. Cancer-related micropeptides encoded by ncRNAs: promising drug targets and prognostic biomarkers. Cancer Lett. 547, 215723 (2022).
He, L. et al. A microRNA polycistron as a potential human oncogene. Nature 435, 828–833 (2005).
Davalos, V. et al. Dynamic epigenetic regulation of the microRNA-200 family mediates epithelial and mesenchymal transitions in human tumorigenesis. Oncogene 31, 2062–2074 (2012).
Toyota, M. et al. Epigenetic silencing of microRNA-34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 68, 4123–4132 (2008).
Kumar, M. S., Lu, J., Mercer, K. L., Golub, T. R. & Jacks, T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat. Genet. 39, 673–677 (2007).
Kawahara, Y. et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315, 1137–1140 (2007).
Bayraktar, R. et al. The mutational landscape and functional effects of noncoding ultraconserved elements in human cancers. Sci. Adv. 11, eado2830 (2025).
Anfossi, S., Babayan, A., Pantel, K. & Calin, G. A. Clinical utility of circulating non-coding RNAs — an update. Nat. Rev. Clin. Oncol. 15, 541–563 (2018).
Dragomir, M. P., Kopetz, S., Ajani, J. A. & Calin, G. A. Non-coding RNAs in GI cancers: from cancer hallmarks to clinical utility. Gut 69, 748–763 (2020).
Matsuzaki, J. et al. Prediction of tissue-of-origin of early stage cancers using serum miRNomes. JNCI Cancer Spectr. 7, pkac080 (2023).
Hogdall, D. et al. Whole blood microRNAs capture systemic reprogramming and have diagnostic potential in patients with biliary tract cancer. J. Hepatol. 77, 1047–1058 (2022).
Miyoshi, J. et al. Liquid biopsy to identify Barrett’s oesophagus, dysplasia and oesophageal adenocarcinoma: the EMERALD multicentre study. Gut 74, 169–181 (2025).
Pardini, B. et al. A fecal microRNA signature by small RNA sequencing accurately distinguishes colorectal cancers: results from a multicenter study. Gastroenterology 165, 582–599.e8 (2023).
Naccarati, A. et al. Fecal miRNA profiles in colorectal cancers with mucinous morphology. Mutagenesis 40, 71–79 (2025).
Wu, P. et al. Pan-cancer characterization of cell-free immune-related miRNA identified as a robust biomarker for cancer diagnosis. Mol. Cancer 23, 31 (2024).
Metcalf, G. A. D. MicroRNAs: circulating biomarkers for the early detection of imperceptible cancers via biosensor and machine-learning advances. Oncogene 43, 2135–2142 (2024).
Ko, S. Y. et al. The glycoprotein CD147 defines miRNA-enriched extracellular vesicles that derive from cancer cells. J. Extracell. Vesicles 12, e12318 (2023).
Li, B. et al. Five miRNAs identified in fucosylated extracellular vesicles as non-invasive diagnostic signatures for hepatocellular carcinoma. Cell Rep. Med. 5, 101716 (2024).
Pinho, S. S. & Reis, C. A. Glycosylation in cancer: mechanisms and clinical implications. Nat. Rev. Cancer 15, 540–555 (2015).
Mitchell, M. I. et al. Exhaled breath condensate contains extracellular vesicles (EVs) that carry miRNA cargos of lung tissue origin that can be selectively purified and analyzed. J. Extracell. Vesicles 13, e12440 (2024).
Li, K. et al. Salivary extracellular microRNAs for early detection and prognostication of esophageal cancer: a clinical study. Gastroenterology 165, 932–945.e9 (2023).
Lei, Y. et al. Simultaneous subset tracing and miRNA profiling of tumor-derived exosomes via dual-surface-protein orthogonal barcoding. Sci. Adv. 9, eadi1556 (2023).
Deamer, D., Akeson, M. & Branton, D. Three decades of nanopore sequencing. Nat. Biotechnol. 34, 518–524 (2016).
Branton, D. et al. The potential and challenges of nanopore sequencing. Nat. Biotechnol. 26, 1146–1153 (2008).
Cai, S. et al. Single-molecule amplification-free multiplexed detection of circulating microRNA cancer biomarkers from serum. Nat. Commun. 12, 3515 (2021).
Zhou, J. et al. High-throughput single-EV liquid biopsy: rapid, simultaneous, and multiplexed detection of nucleic acids, proteins, and their combinations. Sci. Adv. 6, eabc1204 (2020).
Li, Z. et al. Colocalization of protein and microRNA markers reveals unique extracellular vesicle subpopulations for early cancer detection. Sci. Adv. 10, eadh8689 (2024).
Moisoiu, T. et al. Combined miRNA and SERS urine liquid biopsy for the point-of-care diagnosis and molecular stratification of bladder cancer. Mol. Med. 28, 39 (2022).
Wu, Y. et al. DNA-methylation signature accurately differentiates pancreatic cancer from chronic pancreatitis in tissue and plasma. Gut 72, 2344–2353 (2023).
Pastorino, U. et al. Baseline computed tomography screening and blood microRNA predict lung cancer risk and define adequate intervals in the BioMILD trial. Ann. Oncol. 33, 395–405 (2022).
Nappi, L. et al. Integrated expression of circulating miR375 and miR371 to identify teratoma and active germ cell malignancy components in malignant germ cell tumors. Eur. Urol. 79, 16–19 (2021).
Li, X. et al. A predictive system comprising serum microRNAs and radiomics for residual retroperitoneal masses in metastatic nonseminomatous germ cell tumors. Cell Rep. Med. 5, 101843 (2024).
Kim, Y. et al. Tumor-derived EV miRNA signatures surpass total EV miRNA in supplementing mammography for precision breast cancer diagnosis. Theranostics 14, 6587–6604 (2024).
Bhinder, B., Gilvary, C., Madhukar, N. S. & Elemento, O. Artificial intelligence in cancer research and precision medicine. Cancer Discov. 11, 900–915 (2021).
Lotter, W. et al. Artificial intelligence in oncology: current landscape, challenges, and future directions. Cancer Discov. 14, 711–726 (2024).
Zhu, W. & Kan, X. Neural network cascade optimizes microRNA biomarker selection for nasopharyngeal cancer prognosis. PLoS ONE 9, e110537 (2014).
Luna-Aguirre, C. M. et al. Circulating microRNA expression profile in B-cell acute lymphoblastic leukemia. Cancer Biomark 15, 299–310 (2015).
Kong, D., Wang, K., Zhang, Q.-N. & Bing, Z.-T. Systematic analysis reveals key microRNAs as diagnostic and prognostic factors in progressive stages of lung cancer. Preprint at arXiv https://doi.org/10.48550/arXiv.2201.05408 (2022).
Cimmino, A. et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl Acad. Sci. USA 102, 13944–13949 (2005).
Hosseiniyan Khatibi, S. M., Ardalan, M., Teshnehlab, M., Vahed, S. Z. & Pirmoradi, S. Panels of mRNAs and miRNAs for decoding molecular mechanisms of renal cell carcinoma (RCC) subtypes utilizing artificial intelligence approaches. Sci. Rep. 12, 16393 (2022).
Katipally, R. R. et al. Integrated clinical-molecular classification of colorectal liver metastases: a biomarker analysis of the phase 3 new EPOC randomized clinical trial. JAMA Oncol. 9, 1245–1254 (2023).
Keller, A. et al. Competitive learning suggests circulating miRNA profiles for cancers decades prior to diagnosis. RNA Biol. 17, 1416–1426 (2020).
Li, J., Zhang, H. & Gao, F. Identification of miRNA biomarkers for breast cancer by combining ensemble regularized multinomial logistic regression and cox regression. BMC Bioinforma. 23, 434 (2022).
Sarkar, J. P., Saha, I., Sarkar, A. & Maulik, U. Machine learning integrated ensemble of feature selection methods followed by survival analysis for predicting breast cancer subtype specific miRNA biomarkers. Comput. Biol. Med. 131, 104244 (2021).
Zhang, J., Rui, H. & Hu, H. Noninvasive multi-cancer detection using blood-based cell-free microRNAs. Sci. Rep. 14, 22136 (2024).
Kamkar, L., Saberi, S., Totonchi, M. & Kavousi, K. Circulating microRNA panels for multi-cancer detection and gastric cancer screening: leveraging a network biology approach. BMC Med. Genomics 18, 27 (2025).
Chen, D. et al. Large language models in oncology: a review. BMJ Oncol. 4, e000759 (2025).
Dong, B., Sun, W., Xu, D., Wang, G. & Zhang, T. MDformer: a transformer-based method for predicting miRNA-Disease associations using multi-source feature fusion and maximal meta-path instances encoding. Comput. Biol. Med. 167, 107585 (2023).
Waqas, A. et al. Self-normalizing foundation model for enhanced multi-omics data analysis in oncology. Preprint at arXiv https://doi.org/10.48550/arXiv.2405.08226 (2024).
Angeles, A. et al. Matching plasma and tissue miRNA expression analysis to detect viable ovarian germ cell tumors. PLoS ONE 20, e0322477 (2025).
Sun, F. et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 582, 1564–1568 (2008).
Zhao, Y. & Wang, X. miR-34a targets BCL-2 to suppress the migration and invasion of sinonasal squamous cell carcinoma. Oncol. Lett. 16, 6566–6572 (2018).
Cortez, M. A. et al. PDL1 regulation by p53 via miR-34. J. Natl Cancer Inst. 108, djv303 (2016).
Adams, B. D., Parsons, C. & Slack, F. J. The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas. Expert. Opin. Ther. Targets 20, 737–753 (2016).
Beg, M. S. et al. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 35, 180–188 (2017).
Hong, D. S. et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 122, 1630–1637 (2020).
Telford, B. J. et al. Multi-modal effects of 1B3, a novel synthetic miR-193a-3p mimic, support strong potential for therapeutic intervention in oncology. Oncotarget 12, 422–439 (2021).
Issa, H. et al. Preclinical testing of miRNA-193b-3p mimic in acute myeloid leukemias. Leukemia 37, 1583–1587 (2023).
van den Bosch, M. T. J. et al. Transcriptome-wide analysis reveals insight into tumor suppressor functions of 1B3, a novel synthetic miR-193a-3p mimic. Mol. Ther. Nucleic Acids 23, 1161–1171 (2021).
Kim, T. & Croce, C. M. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp. Mol. Med. 55, 1314–1321 (2023).
Duurland, C. L. et al. INT-1B3, an LNP formulated miR-193a-3p mimic, promotes anti-tumor immunity by enhancing T cell mediated immune responses via modulation of the tumor microenvironment and induction of immunogenic cell death. Oncotarget 15, 470–485 (2024).
van den Bosch, M. T. J. et al. The tumor suppressor 5A2, a synthetic miR-7-5p mimic, targets oncogenic and metabolic pathways, as revealed by transcriptome-wide analysis. Front. Drug Discov. 3, 1181637 (2023).
Seto, A. G. et al. Cobomarsen, an oligonucleotide inhibitor of miR-155, co-ordinately regulates multiple survival pathways to reduce cellular proliferation and survival in cutaneous T-cell lymphoma. Br. J. Haematol. 183, 428–444 (2018).
Sun, X., Setrerrahmane, S., Li, C., Hu, J. & Xu, H. Nucleic acid drugs: recent progress and future perspectives. Signal. Transduct. Target. Ther. 9, 316 (2024).
He, X. et al. Non-coding RNAs in the spotlight of the pathogenesis, diagnosis, and therapy of cutaneous T cell lymphoma. Cell Death Discov. 10, 400 (2024).
Querfeld, C. et al. Phase 1 trial of cobomarsen, an inhibitor of Mir-155, in cutaneous T cell lymphoma [abstract]. Blood 132, 2903 (2018).
Taubel, J. et al. Novel antisense therapy targeting microRNA-132 in patients with heart failure: results of a first-in-human phase 1b randomized, double-blind, placebo-controlled study. Eur. Heart J. 42, 178–188 (2021).
Jdiaa, S. S., Mustafa, R. A. & Yu, A. S. L. Treatment of autosomal-dominant polycystic kidney disease. Am. J. Kidney Dis. 85, 491–500 (2025).
Badanina, D. M. et al. Therapeutic potential of modulating gene-microRNA crosstalk in burn injury. Int. J. Mol. Sci. 26, 10060 (2025).
Bartel, D. P. Metazoan microRNAs. Cell 173, 20–51 (2018).
Yang, N. et al. MicroRNAs: pleiotropic regulators in the tumor microenvironment. Front. Immunol. 9, 2491 (2018).
Jovelin, R. Pleiotropic constraints, expression level, and the evolution of miRNA sequences. J. Mol. Evol. 77, 206–220 (2013).
Ebert, M. S. & Sharp, P. A. Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 (2012).
Lehmann, S. M. et al. An unconventional role for miRNA: let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835 (2012).
Feng, Y. et al. Extracellular microRNAs induce potent innate immune responses via TLR7/MyD88-dependent mechanisms. J. Immunol. 199, 2106–2117 (2017).
Rasko, J. E. & Wong, J. J. Nuclear microRNAs in normal hemopoiesis and cancer. J. Hematol. Oncol. 10, 8 (2017).
Zhou, H. et al. A peptide encoded by pri-miRNA-31 represses autoimmunity by promoting Treg differentiation. EMBO Rep. 23, e53475 (2022).
Mestdagh, P. et al. Evaluation of quantitative miRNA expression platforms in the microRNA quality control (miRQC) study. Nat. Methods 11, 809–815 (2014).
Zelli, V. et al. Emerging role of isomiRs in cancer: state of the art and recent advances. Genes 12, 1447 (2021).
Pinzon, N. et al. microRNA target prediction programs predict many false positives. Genome Res. 27, 234–245 (2017).
Baek, D. et al. The impact of microRNAs on protein output. Nature 455, 64–71 (2008).
Chi, S. W., Zang, J. B., Mele, A. & Darnell, R. B. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460, 479–486 (2009).
Hum, N. R. et al. Comparative molecular analysis of cancer behavior cultured in vitro, in vivo, and ex vivo. Cancers 12, 690 (2020).
Kanasty, R., Dorkin, J. R., Vegas, A. & Anderson, D. Delivery materials for siRNA therapeutics. Nat. Mater. 12, 967–977 (2013).
Almarza, E. et al. Characteristics of lentiviral vectors harboring the proximal promoter of the vav proto-oncogene: a weak and efficient promoter for gene therapy. Mol. Ther. 15, 1487–1494 (2007).
Wittrup, A. & Lieberman, J. Knocking down disease: a progress report on siRNA therapeutics. Nat. Rev. Genet. 16, 543–552 (2015).
McDonald, J. S., Milosevic, D., Reddi, H. V., Grebe, S. K. & Algeciras-Schimnich, A. Analysis of circulating microRNA: preanalytical and analytical challenges. Clin. Chem. 57, 833–840 (2011).
Leidinger, P. et al. A blood based 12-miRNA signature of Alzheimer disease patients. Genome Biol. 14, R78 (2013).
Libbrecht, M. W. & Noble, W. S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 16, 321–332 (2015).
van Rooij, E. & Kauppinen, S. Development of microRNA therapeutics is coming of age. EMBO Mol. Med. 6, 851–864 (2014).
Hafner, M. et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141, 129–141 (2010).
Ingolia, N. T., Ghaemmaghami, S., Newman, J. R. & Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223 (2009).
Faridani, O. R. et al. Single-cell sequencing of the small-RNA transcriptome. Nat. Biotechnol. 34, 1264–1266 (2016).
McKellar, D. W. et al. Spatial mapping of the total transcriptome by in situ polyadenylation. Nat. Biotechnol. 41, 513–520 (2023).
Su, J. et al. Cell–cell communication: new insights and clinical implications. Signal. Transduct. Target. Ther. 9, 196 (2024).
Ijee, S. et al. Efficient deletion of microRNAs using CRISPR/Cas9 with dual guide RNAs. Front. Mol. Biosci. 10, 1295507 (2023).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Zafra, M. P. et al. Optimized base editors enable efficient editing in cells, organoids and mice. Nat. Biotechnol. 36, 888–893 (2018).
Klemke, M. et al. Loss of let-7 binding sites resulting from truncations of the 3′ untranslated region of HMGA2 mRNA in uterine leiomyomas. Cancer Genet. Cytogenet. 196, 119–123 (2010).
Chen, K. et al. The overlooked fact: fundamental need for spike-in control for virtually all genome-wide analyses. Mol. Cell Biol. 36, 662–667 (2015).
ElHefnawi, M., Soliman, B., Abu-Shahba, N. & Amer, M. An integrative meta-analysis of microRNAs in hepatocellular carcinoma. Genomics Proteom. Bioinforma. 11, 354–367 (2013).
Fu, L. & Peng, Q. A deep ensemble model to predict miRNA–disease association. Sci. Rep. 7, 14482 (2017).
Zhang, Y., Parmigiani, G. & Johnson, W. E. ComBat-seq: batch effect adjustment for RNA-seq count data. NAR. Genom. Bioinform 2, lqaa078 (2020).
Leek, J. T., Johnson, W. E., Parker, H. S., Jaffe, A. E. & Storey, J. D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883 (2012).
Sun, Y. & Qiu, P. Domain adaptation for supervised integration of scRNA-seq data. Commun. Biol. 6, 274 (2023).
Wagner, V., Meese, E. & Keller, A. The intricacies of isomiRs: from classification to clinical relevance. Trends Genet. 40, 784–796 (2024).
Pla, A., Zhong, X. & Rayner, S. miRAW: a deep learning-based approach to predict microRNA targets by analyzing whole microRNA transcripts. PLoS Comput. Biol. 14, e1006185 (2018).
Gu, T., Zhao, X., Barbazuk, W. B. & Lee, J. H. miTAR: a hybrid deep learning-based approach for predicting miRNA targets. BMC Bioinforma. 22, 96 (2021).
Dalla-Torre, H. et al. Nucleotide Transformer: building and evaluating robust foundation models for human genomics. Nat. Methods 22, 287–297 (2025).
Xie, X., Wang, Y., He, K. & Sheng, N. Predicting miRNA-disease associations based on PPMI and attention network. BMC Bioinforma. 24, 113 (2023).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Zuo, L. et al. Exosomes-coated miR-34a displays potent antitumor activity in pancreatic cancer both in vitro and in vivo. Drug Des. Devel. Ther. 14, 3495–3507 (2020).
Paliwal, S. R., Paliwal, R. & Vyas, S. P. A review of mechanistic insight and application of pH-sensitive liposomes in drug delivery. Drug Deliv. 22, 231–242 (2015).
Yuba, E. Design of pH-sensitive polymer-modified liposomes for antigen delivery and their application in cancer immunotherapy. Polym. J. 48, 761–771 (2016).
Kocak, G., Tuncer, C. & Bücün, V. pH-Responsive polymers. Polym. Chem. 8, 144–176 (2017).
Sun, L. et al. Smart nanoparticles for cancer therapy. Signal. Transduct. Target. Ther. 8, 418 (2023).
Sugahara, K. N. et al. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell 16, 510–520 (2009).
Juang, V., Chang, C. H., Wang, C. S., Wang, H. E. & Lo, Y. L. pH-Responsive PEG-shedding and targeting peptide-modified nanoparticles for dual-delivery of irinotecan and microRNA to enhance tumor-specific therapy. Small 15, e1903296 (2019).
Kim, H. I. et al. Recent advances in extracellular vesicles for therapeutic cargo delivery. Exp. Mol. Med. 56, 836–849 (2024).
Volpini, L., Monaco, F., Santarelli, L., Neuzil, J. & Tomasetti, M. Advances in RNA cancer therapeutics: new insight into exosomes as miRNA delivery. Asp. Mol. Med. 1, 100005 (2023).
EL Andaloussi, S., Mäger, I., Breakefield, X. O. & Wood, M. J. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013).
Ohno, S. et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol. Ther. 21, 185–191 (2013).
Kamerkar, S. et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature 546, 498–503 (2017).
Jurj, A., Fontana, B., Varani, G. & Calin, G. A. Small molecules targeting microRNAs: new opportunities and challenges in precision cancer therapy. Trends Cancer 10, 809–824 (2024).
Tadesse, K. & Benhamou, R. I. Targeting microRNAs with small molecules. Noncoding RNA 10, 17 (2024).
Sidharthan, V. et al. Use of a small molecule microarray screen to identify inhibitors of the catalytic RNA subunit of Methanobrevibacter smithii RNase P. Nucleic Acids Res. 53, gkae1190 (2025).
Sedgwick, A. C. et al. Indicator displacement assays (IDAs): the past, present and future. Chem. Soc. Rev. 50, 9–38 (2021).
Bood, M. et al. Interbase-FRET binding assay for pre-microRNAs. Sci. Rep. 11, 9396 (2021).
Wu, M., Huang, W., Yang, N. & Liu, Y. Learn from antibody–drug conjugates: consideration in the future construction of peptide–drug conjugates for cancer therapy. Exp. Hematol. Oncol. 11, 93 (2022).
Meng, Q. et al. Antibody-oligonucleotide conjugates in cancer therapy: potential and promise. Crit. Rev. Oncology/Hematology 215, 104858 (2025).
Song, E. et al. Antibody mediated in vivo delivery of small interfering RNAs via cell-surface receptors. Nat. Biotechnol. 23, 709–717 (2005).
Kashyap, D. & Booth, M. J. Nucleic acid conjugates: unlocking therapeutic potential. ACS Bio Med Chem Au 5, 3–15 (2025).
Cancer Genome Atlas Research Network The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Clough, E. & Barrett, T. The Gene Expression Omnibus database. Methods Mol. Biol. 1418, 93–110 (2016).
Cui, S. et al. miRTarBase 2025: updates to the collection of experimentally validated microRNA-target interactions. Nucleic Acids Res. 53, D147–D156 (2025).
Li, R. et al. CancerMIRNome: an interactive analysis and visualization database for miRNome profiles of human cancer. Nucleic Acids Res. 50, D1139–D1146 (2022).
de Gonzalo-Calvo, D. et al. Machine learning for catalysing the integration of noncoding RNA in research and clinical practice. EBioMedicine 106, 105247 (2024).
Qi, B., Fiori, L. M., Turecki, G. & Trakadis, Y. J. Machine learning analysis of blood microRNA data in major depression: a case-control study for biomarker discovery. Int. J. Neuropsychopharmacol. 23, 505–510 (2020).
Zhang, Y. et al. Machine learning-based exceptional response prediction of nivolumab monotherapy with circulating microRNAs in non-small cell lung cancer. Lung Cancer 173, 107–115 (2022).
Fehlmann, T. et al. Evaluating the use of circulating microRNA profiles for lung cancer detection in symptomatic patients. JAMA Oncol. 6, 714–723 (2020).
Wang, Y., Carter, B. Z., Li, Z. & Huang, X. Application of machine learning methods in clinical trials for precision medicine. JAMIA Open. 5, ooab107 (2022).
Zhang, B. et al. Harnessing artificial intelligence to improve clinical trial design. Commun. Med. 3, 191 (2023).
Kavalci, E. & Hartshorn, A. Improving clinical trial design using interpretable machine learning based prediction of early trial termination. Sci. Rep. 13, 121 (2023).
Franke, J. K., Runge, F., Köksal, R., Backofen, R. & Hutter, F. RNAformer: a simple yet effective deep learning model for RNA secondary structure prediction. Preprint at bioRxiv https://doi.org/10.1101/2024.02.12.579881 (2024).
Li, F. et al. Advancing mRNA subcellular localization prediction with graph neural network and RNA structure. Bioinformatics 40, btae504 (2024).
Karimi, M., Wu, D., Wang, Z. & Shen, Y. DeepAffinity: interpretable deep learning of compound-protein affinity through unified recurrent and convolutional neural networks. Bioinformatics 35, 3329–3338 (2019).
Krishnan, S. R., Roy, A. & Gromiha, M. M. Reliable method for predicting the binding affinity of RNA-small molecule interactions using machine learning. Brief. Bioinform. 25, bbae002 (2024).
Alipanahi, B., Delong, A., Weirauch, M. T. & Frey, B. J. Predicting the sequence specificities of DNA- and RNA-binding proteins by deep learning. Nat. Biotechnol. 33, 831–838 (2015).
Sun, S. & Gao, L. Contrastive pre-training and 3D convolution neural network for RNA and small molecule binding affinity prediction. Bioinformatics 40, btae155 (2024).
Galeano, D. et al. sChemNET: a deep learning framework for predicting small molecules targeting microRNA function. Nat. Commun. 15, 9149 (2024).
Tong, Y. et al. Programming inactive RNA-binding small molecules into bioactive degraders. Nature 618, 169–179 (2023).
Dietterich, T. Overfitting and undercomputing in machine learning. ACM Comput. Surv. 27, 326–327 (1995).
Shi, W. et al. Integrating a microRNA signature as a liquid biopsy-based tool for the early diagnosis and prediction of potential therapeutic targets in pancreatic cancer. Br. J. Cancer 130, 125–134 (2024).
Hamidi, F. et al. Identifying potential circulating miRNA biomarkers for the diagnosis and prediction of ovarian cancer using machine-learning approach: application of Boruta. Front. Digit. Health 5, 1187578 (2023).
Guidotti, R. et al. A survey of methods for explaining black box models. ACM Comput. Surv. 51, 93 (2018).
Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 1, 206–215 (2019).
Dwivedi, R. et al. Explainable AI (XAI): core ideas, techniques, and solutions. ACM Comput. Surv. 55, 194 (2023).
Lu, J. et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838 (2005).
Chafik, A. Machine learning-based selection of key miRNA biomarkers for breast cancer diagnostics. J. Appl. Sci. 15, 597–603 (2025).
Yerukala Sathipati, S., Tsai, M. J., Shukla, S. K. & Ho, S. Y. Artificial intelligence-driven pan-cancer analysis reveals miRNA signatures for cancer stage prediction. HGG Adv. 4, 100190 (2023).
Singh, S. et al. Integrating miRNA profiling and machine learning for improved prostate cancer diagnosis. Sci. Rep. 15, 30477 (2025).
Pawelka, D. et al. Machine-learning-based analysis identifies miRNA expression profile for diagnosis and prediction of colorectal cancer: a preliminary study. Cancer Genomics Proteom. 19, 503–511 (2022).
Sathipati, S. Y. et al. An evolutionary learning-based method for identifying a circulating miRNA signature for breast cancer diagnosis prediction. NAR. Genom. Bioinform 6, lqae022 (2024).
Azari, H. et al. Machine learning algorithms reveal potential miRNAs biomarkers in gastric cancer. Sci. Rep. 13, 6147 (2023).
Kantamnen, S., Hegde, P., Patil, N. Utilizing deep learning methods for cancer detection through analysis of microRNA expression profiles. 2024 15th International Conference on Computing Communication and Networking Technologies (ICCCNT) https://doi.org/10.1109/ICCCNT61001.2024.10725601 (IEEE, 2024).
Pal, J. K. & Rami, B. R. Machine learning based identification of candidate miRNA biomarkers for micro-invasive breast cancer diagnosis. Preprint at bioRxiv https://doi.org/10.1101/2024.08.16.608025 (2024).
Alizadeh Savareh, B. et al. A machine learning approach identified a diagnostic model for pancreatic cancer through using circulating microRNA signatures. Pancreatology 20, 1195–1204 (2020).
Sarkar, A., Das, T., Das, G. & Ghosh, Z. MicroRNA mediated gene regulatory circuits leads to machine learning based preliminary detection of acute myeloid leukemia. Comput. Biol. Chem. 104, 107859 (2023).
Wang, C., Kouznetsova, V. L., Kesari, S. & Tsigelny, I. F. Developing a novel medulloblastoma diagnostic with miRNA biomarkers and machine learning. Childs Nerv. Syst. 41, 221 (2025).
Tassone, P. et al. Safety and activity of the first-in-class locked nucleic acid (LNA) miR-221 selective inhibitor in refractory advanced cancer patients: a first-in-human, phase 1, open-label, dose-escalation study. J. Hematol. Oncol. 16, 68 (2023).
van Zandwijk, N. et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: a first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 18, 1386–1396 (2017).
Haniff, H. S. et al. Design of a small molecule that stimulates vascular endothelial growth factor A enabled by screening RNA fold-small molecule interactions. Nat. Chem. 12, 952–961 (2020).
Chang, L., Zhou, G., Soufan, O. & Xia, J. miRNet 2.0: network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 48, W244–W251 (2020).
Wen, M., Cong, P., Zhang, Z., Lu, H. & Li, T. DeepMirTar: a deep-learning approach for predicting human miRNA targets. Bioinformatics 34, 3781–3787 (2018).
Lee, B., Baek, J., Park, J., Yoon, S. deepTarget: end-to-end learning framework for microRNA target prediction using deep recurrent neural networks. BCB '16: Proceedings of the 7th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics 434–442 (Association for Computing Machinery, 2016).
Hitzler, P., Eberhart, A., Ebrahimi, M., Sarker, M. K. & Zhou, L. Neuro-symbolic approaches in artificial intelligence. Natl Sci. Rev. 9, nwac035 (2022).
Coronato, A., Naeem, M., De Pietro, G. & Paragliola, G. Reinforcement learning for intelligent healthcare applications: a survey. Artif. Intell. Med. 109, 101964 (2020).
Harshvardhan, G. M., Mahendra, K. G., Manjusha, P. & Siddharth, S. R. A comprehensive survey and analysis of generative models in machine learning. Computer Sci. Rev. 38, 100285 (2020).
Noble, W. S. What is a support vector machine? Nat. Biotechnol. 24, 1565–1567 (2006).
Golub, T. R. et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 286, 531–537 (1999).
Krizhevsky, A., Sutskever, I. & Hinton, G. E. ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems vol. 25 (eds Pereira, F., Burges, C. J., Bottou, L. & Weinberger, K. Q.) (Curran Associates, 2012).
Ehteshami Bejnordi, B. et al. Using deep convolutional neural networks to identify and classify tumor-associated stroma in diagnostic breast biopsies. Mod. Pathol. 31, 1502–1512 (2018).
Esteva, A. et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 542, 115–118 (2017).
Castro-Vega, L. J. et al. Multi-omics analysis defines core genomic alterations in pheochromocytomas and paragangliomas. Nat. Commun. 6, 6044 (2015).
Esteva, A. et al. A guide to deep learning in healthcare. Nat. Med. 25, 24–29 (2019).
Yang, X. et al. A large language model for electronic health records. NPJ Digit. Med. 5, 194 (2022).
Pai, S. et al. Foundation model for cancer imaging biomarkers. Nat. Mach. Intell. 6, 354–367 (2024).
Vorontsov, E. et al. A foundation model for clinical-grade computational pathology and rare cancers detection. Nat. Med. 30, 2924–2935 (2024).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Hearst, M. A., Dumais, S. T., Osuna, E., Platt, J. & Scholkopf, B. Support vector machines. IEEE Intell. Syst. their Appl. 13, 18–28 (1998).
Gurney, K. An Introduction to Neural Networks (CRC Press, 2018).
Huang, Y. et al. Spatial dissection of invasive front from tumor mass enables discovery of novel microRNA drivers of glioblastoma invasion. Adv. Sci. 8, e2101923 (2021).
Giubellino, A., Ricketts, C. J., Moreno, V., Linehan, W. M. & Merino, M. J. MicroRNA profiling of morphologically heterogeneous clear cell renal cell carcinoma. J. Cancer 12, 5375–5384 (2021).
Statello, L., Guo, C. J., Chen, L. L. & Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 22, 96–118 (2021).
Isakova, A., Fehlmann, T., Keller, A. & Quake, S. R. A mouse tissue atlas of small noncoding RNA. Proc. Natl Acad. Sci. USA 117, 25634–25645 (2020).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Stahl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Junker, J. P. et al. Genome-wide RNA tomography in the zebrafish embryo. Cell 159, 662–675 (2014).
Jayaprakash, A. D., Jabado, O., Brown, B. D. & Sachidanandam, R. Identification and remediation of biases in the activity of RNA ligases in small-RNA deep sequencing. Nucleic Acids Res. 39, e141 (2011).
Maji, R. K., Leisegang, M. S., Boon, R. A. & Schulz, M. H. Revealing microRNA regulation in single cells. Trends Genet. 41, 522–536 (2025).
Linsen, S. E. et al. Limitations and possibilities of small RNA digital gene expression profiling. Nat. Methods 6, 474–476 (2009).
Shore, S. et al. Small RNA library preparation method for next-generation sequencing using chemical modifications to prevent adapter dimer formation. PLoS ONE 11, e0167009 (2016).
Hucker, S. M. et al. Single-cell microRNA sequencing method comparison and application to cell lines and circulating lung tumor cells. Nat. Commun. 12, 4316 (2021).
Xiao, Z. et al. Holo-Seq: single-cell sequencing of holo-transcriptome. Genome Biol. 19, 163 (2018).
Wang, N. et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat. Commun. 10, 95 (2019).
Isakova, A., Neff, N. & Quake, S. R. Single-cell quantification of a broad RNA spectrum reveals unique noncoding patterns associated with cell types and states. Proc. Natl Acad. Sci. USA 118, e2113568118 (2021).
Bai, Z. et al. Spatially exploring RNA biology in archival formalin-fixed paraffin-embedded tissues. Cell 187, 6760–6779.e24 (2024).
Piwecka, M. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017).
Acknowledgements
G.A.C. is the Charles B. Barker Chair. Work in G.A.C.’s laboratory is supported by NCI grant 1R01CA222007-01A1, NIDCR grant R01DE032018, DoD CDRMP Idea Award BC200208P1 (W81XWH-21-1-0030), Team DoD Grant in Gastric Cancer CA200990P2 (W81XWH-21-1-0715), DoD Idea Award PC230419 (HT9425-24-1-0052), the 2019 Faculty Achievement Award, CLL Global Research Foundation 2019 grant, CLL Global Research Foundation 2020 grant, CLL Global Research Foundation 2022 grant, CLL Global Research Foundation 2024 grant, The G. Harold & Leila Y. Mathers Foundation, two grants from Torrey Coast Foundation, an Institutional Research Grant 2024, a Development Grant associated with the Brain SPORE 2P50CA127001, an Institutional Bridge Funding grant 2023, and the Ben and Catherine Ivy Foundation grant. The work of Z.L. was partially supported by US NIGMS grant R35GM159819 and a Translational and Basic Science Research in Early Lesions (TBEL) Coordinating and Data Management Center (CDMC) grant U24CA274212. The work of M.P.D. is supported by the Berlin Institute of Health Clinician Scientist Program, a German Cancer Research Center (DKTK) Berlin Young Investigator Grant 2022, the Berliner Krebsgesellschaft DRFF202204, and an Else Kröner-Fresenius Foundation First and Second Applications Grant (2024_EKEA.77).
Author information
Authors and Affiliations
Contributions
All authors made a substantial contribution to all aspects of the preparation of this manuscript.
Corresponding authors
Ethics declarations
Competing interests
G.A.C. is one of the scientific founders of Ithax Therapeutics. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Clinical Oncology thanks H.-D. Huang, N. Seki and the other, anonymous, reviewer for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Adaptor dimer
-
Sequencing phenomenon whereby two adaptors bind to each other or build short adaptor-only fragments with consequences such as wasted sequencing reads and/or reduced depth per-cell depth, especially problematic when cDNA amounts are low.
- Adaptor ligation bias
-
Certain RNA sequences are favoured during adaptor ligation, which can distort gene expression quantification and result in cell type-specific artefacts.
- Artificial intelligence
-
A broad field focused on building computational systems that mimic human intelligence, including reasoning, learning and decision-making.
- Black box
-
A term used to describe artificial intelligence and machine learning models with internal decision-making processes that are difficult to interpret, with a lack of transparency regarding the biological rationale underlying their predictions that might reduce trust in the outputs, thus posing challenges for clinical translation.
- Decision trees
-
A supervised machine learning method that uses a tree-like structure to make predictions. At each branching point (‘node’), the algorithm applies a decision rule based on one feature, splitting the data into subgroups. This process continues until terminal nodes (‘leaves’), which assign a final prediction, are reached. Decision trees are highly interpretable and form the basis for many ensemble approaches such as random forests and boosting algorithms.
- Ensemble learning
-
A machine learning strategy in which multiple predictive models (for example, decision trees, neural networks or statistical classifiers) are combined to improve overall performance.
- Feature spaces
-
Multidimensional spaces in which each dimension represents a measurable attribute or feature of the data. Each sample can be visualized as a point in such a space, and the geometric relationships between points often reflect underlying biological or clinical differences. Machine learning models operate within these feature spaces to detect patterns, classify samples, or make predictions.
- Hyperplane
-
A hyperplane is a mathematical boundary that separates data points into different groups within a multidimensional feature space. In machine learning, classifiers such as support vector machines use hyperplanes to distinguish between classes by finding the boundary that best maximizes the margin between them.
- Kernel tricks
-
A computational technique used in algorithms such as support vector machines to analyse data that are not linearly separable. The kernel trick maps the original data into a higher-dimensional feature space whereby a simple linear boundary (hyperplane) can separate the classes without explicitly computing the high-dimensional transformation. This approach enables efficient modelling of complex, non-linear relationships.
- Locked nucleic acid
-
(LNA). A chemically modified nucleic acid analogue in which the ribose sugar is conformationally constrained by a 2′-O,4′-C methylene bridge. This ‘locked’ C3′ endo conformation increases the thermal stability and affinity of LNA-containing oligonucleotides for complementary RNA, enhances mismatch discrimination, and provides strong resistance to nuclease degradation. LNAs are widely used in antisense and microRNA-based therapeutics to improve potency, specificity and in vivo stability.
- Machine learning
-
A key form of artificial intelligence in which data are used to train computational models to identify patterns and make predictions or decisions without explicit programming.
- miRNA signatures
-
Defined sets of microRNAs (miRNAs), the combined expression pattern of which distinguishes biological states such as cancer from non-malignant tissue and tumour subtypes as well as disease prognosis or responsiveness to therapy. Unlike single miRNA species, miRNA signatures capture the network-level regulation of gene expression and often provide greater sensitivity, specificity and robustness in clinical applications.
- miRNome
-
The complete set of microRNAs (miRNAs) expressed in a specific cell type, tissue or organism under defined conditions. Analysis of the miRNome enables comprehensive profiling of miRNA expression patterns, identification of disease-associated miRNA signatures and the study of regulatory networks involving miRNAs.
- Seed matching
-
Putative microRNA (miRNA) targets are identified by complementarity within the seed region (nucleotides 2–7/8 of the miRNA). Although seed pairing is the primary determinant of target specificity, it often yields false-positive results because short complementary motifs occur by chance, and functional targeting also requires favourable mRNA structure, site accessibility, sequence context and co-expression of the miRNA and its target.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jurj, A., Dragomir, M.P., Li, Z. et al. MicroRNAs in oncology: a translational perspective in the era of AI. Nat Rev Clin Oncol (2026). https://doi.org/10.1038/s41571-025-01114-x
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41571-025-01114-x