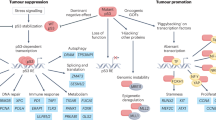
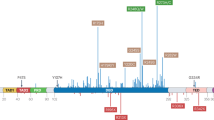
Abstract
Mutations in the TP53 tumour suppressor gene are very frequent in cancer, and attempts to restore the functionality of p53 in tumours as a therapeutic strategy began decades ago. However, very few of these drug development programmes have reached late-stage clinical trials, and no p53-based therapeutics have been approved in the USA or Europe so far. This is probably because, as a nuclear transcription factor, p53 does not possess typical drug target features and has therefore long been considered undruggable. Nevertheless, several promising approaches towards p53-based therapy have emerged in recent years, including improved versions of earlier strategies and novel approaches to make undruggable targets druggable. Small molecules that can either protect p53 from its negative regulators or restore the functionality of mutant p53 proteins are gaining interest, and drugs tailored to specific types of p53 mutants are emerging. In parallel, there is renewed interest in gene therapy strategies and p53-based immunotherapy approaches. However, major concerns still remain to be addressed. This Review re-evaluates the efforts made towards targeting p53-dysfunctional cancers, and discusses the challenges encountered during clinical development.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Barbosa, A. et al. Gene panel tumor testing in ovarian cancer patients significantly increases the yield of clinically actionable germline variants beyond BRCA1/BRCA2. Cancers 12, 2834 (2020).
Hoe, K. K., Verma, C. S. & Lane, D. P. Drugging the p53 pathway: understanding the route to clinical efficacy. Nat. Rev. Drug Discov. 13, 217–236 (2014).
Sabapathy, K. & Lane, D. P. Therapeutic targeting of p53: all mutants are equal, but some mutants are more equal than others. Nat. Rev. Clin. Oncol. 15, 13–30 (2018).
Bykov, V. J. N., Eriksson, S. E., Bianchi, J. & Wiman, K. G. Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer 182, 89–102 (2017).
Cheok, C. F. & Lane, D. P. Exploiting the p53 pathway for therapy. Cold Spring Harb. Perspect. Med. 7, a026310 (2017).
Levine, A. J. p53: 800 million years of evolution and 40 years of discovery. Nat. Rev. Cancer 20, 471–480 (2020).
Kastan, M. B., Canman, C. E. & Leonard, C. J. p53, cell cycle control and apoptosis: implications for cancer. Cancer Metastasis Rev. 14, 3–15 (1995).
Williams, A. B. & Schumacher, B. p53 in the DNA-damage-repair process. Cold Spring Harb. Perspect. Med. 6, a026070 (2016).
Fridman, J. S. & Lowe, S. W. Control of apoptosis by p53. Oncogene 22, 9030–9040 (2003).
Kastenhuber, E. R. & Lowe, S. W. Putting p53 in context. Cell 170, 1062–1078 (2017).
Speidel, D. Transcription-independent p53 apoptosis: an alternative route to death. Trends Cell Biol. 20, 14–24 (2010).
Vaseva, A. V. & Moll, U. M. The mitochondrial p53 pathway. Biochim. Biophys. Acta 1787, 414 (2009).
Giorgi, C. et al. p53 at the endoplasmic reticulum regulates apoptosis in a Ca2+-dependent manner. Proc. Natl Acad. Sci. USA 112, 1779–1784 (2015).
Maiuri, M. C. et al. Autophagy regulation by p53. Curr. Opin. Cell Biol. 22, 181–185 (2010).
Tan, X. et al. p53 loss activates prometastatic secretory vesicle biogenesis in the Golgi. Sci. Adv. 7, eabf4885 (2021).
Mantovani, F., Collavin, L. & Del Sal, G. Mutant p53 as a guardian of the cancer cell. Cell Death Differ. 26, 199–212 (2018).
Sicari, D. et al. Mutant p53 improves cancer cells’ resistance to endoplasmic reticulum stress by sustaining activation of the UPR regulator ATF6. Oncogene 38, 6184–6195 (2019).
Walerych, D. et al. Proteasome machinery is instrumental in a common gain-of-function program of the p53 missense mutants in cancer. Nat. Cell Biol. 18, 897–909 (2016).
Capaci, V. et al. Mutant p53 induces Golgi tubulo-vesiculation driving a prometastatic secretome. Nat. Commun. 11, 3945 (2020).
Weisz, L., Oren, M. & Rotter, V. Transcription regulation by mutant p53. Oncogene 26, 2202–2211 (2007).
Kim, M. P. & Lozano, G. Mutant p53 partners in crime. Cell Death Differ. 25, 161–168 (2018).
Amit, M. et al. Loss of p53 drives neuron reprogramming in head and neck cancer. Nature 578, 449–454 (2020).
Blagih, J., Buck, M. D. & Vousden, K. H. p53, cancer and the immune response. J. Cell Sci. 133, jcs237453 (2020).
Wellenstein, M. D. et al. Loss of p53 triggers WNT-dependent systemic inflammation to drive breast cancer metastasis. Nature 572, 538–542 (2019). This paper shows that p53 loss in cancer cells promotes a tumour-supportive immune microenvironment in multiple mouse models of breast cancer.
Cooks, T. et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat. Commun. 9, 771 (2018).
Pilley, S., Rodriguez, T. A. & Vousden, K. H. Mutant p53 in cell-cell interactions. Genes Dev. 35, 433–448 (2021).
Olivier, M., Hollstein, M. & Hainaut, P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb. Perspect. Biol. 2, a001008 (2010).
Shchors, K. et al. Using a preclinical mouse model of high-grade astrocytoma to optimize p53 restoration therapy. Proc. Natl Acad. Sci. USA 110, E1480–E1489 (2013).
Martins, C. P., Brown-Swigart, L. & Evan, G. I. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell 127, 1323–1334 (2006).
Xue, W. et al. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature 445, 656–660 (2007).
Ventura, A. et al. Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665 (2007).
Sánchez-Rivera, F. J. et al. Mitochondrial apoptotic priming is a key determinant of cell fate upon p53 restoration. Proc. Natl Acad. Sci. USA 118, e2019740118 (2021).
Foster, B. A., Coffey, H. A., Morin, M. J. & Rastinejad, F. Pharmacological rescue of mutant p53 conformation and function. Science 286, 2507–2510 (1999).
Wang, W., Takimoto, R., Rastinejad, F. & El-Deiry, W. S. Stabilization of p53 by CP-31398 inhibits ubiquitination without altering phosphorylation at serine 15 or 20 or MDM2 binding. Mol. Cell. Biol. 23, 2171–2181 (2003).
Rippin, T. M. et al. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene 21, 2119–2129 (2002).
Bykov, V. J. N. et al. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8, 282–288 (2002). This is the first report of PRIMA-1, a small molecule that restores wtp53-like functionality to multiple p53 mutants and induces apoptosis in cancer cells.
Lambert, J. M. R. et al. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15, 376–388 (2009).
Bykov, V. J. N. et al. Targeting of mutant p53 and the cellular redox balance by APR-246 as a strategy for efficient cancer therapy. Front. Oncol. 6, 21 (2016).
Chipuk, J. E., Maurer, U., Green, D. R. & Schuler, M. Pharmacologic activation of p53 elicits Bax-dependent apoptosis in the absence of transcription. Cancer Cell 4, 371–381 (2003).
Saha, M. N., Chen, Y., Chen, M. H., Chen, G. & Chang, H. Small molecule MIRA-1 induces in vitro and in vivo anti-myeloma activity and synergizes with current anti-myeloma agents. Br. J. Cancer 110, 2224–2231 (2014).
Zache, N. et al. Mutant p53 targeting by the low molecular weight compound STIMA-1. Mol. Oncol. 2, 70–80 (2008).
Bou-Hanna, C. et al. Acute cytotoxicity of MIRA-1/NSC19630, a mutant p53-reactivating small molecule, against human normal and cancer cells via a caspase-9-dependent apoptosis. Cancer Lett. 359, 211–217 (2015).
Tal, P. et al. Cancer therapeutic approach based on conformational stabilization of mutant p53 protein by small peptides. Oncotarget 7, 11817–11837 (2016).
Ali, D. et al. APR-246 exhibits anti-leukemic activity and synergism with conventional chemotherapeutic drugs in acute myeloid leukemia cells. Eur. J. Haematol. 86, 206–215 (2011).
Zandi, R. et al. PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin. Cancer Res. 17, 2830–2841 (2011).
Liu, X. et al. Small molecule induced reactivation of mutant p53 in cancer cells. Nucleic Acids Res. 41, 6034–6044 (2013).
Bykov, V. J. N. et al. PRIMA-1MET synergizes with cisplatin to induce tumor cell apoptosis. Oncogene 24, 3484–3491 (2005).
Mohell, N. et al. APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis. 6, e1794 (2015).
Fransson, Å. et al. Strong synergy with APR-246 and DNA-damaging drugs in primary cancer cells from patients with TP53 mutant high-grade serous ovarian cancer. J. Ovarian Res. 9, 27 (2016).
Sallman, D. A. et al. Eprenetapopt (APR-246) and azacitidine in TP53-mutant myelodysplastic syndromes. J. Clin. Oncol. 39, 1584–1594 (2021).
Cluzeau, T. et al. Eprenetapopt plus azacitidine in TP53-mutated myelodysplastic syndromes and acute myeloid leukemia: a phase II study by the Groupe Francophone des Myélodysplasies (GFM). J. Clin. Oncol. 39, 1575–1583 (2021).
Aprea https://ir.aprea.com/news-releases/news-release-details/aprea-therapeutics-announces-positive-results-phase-2-trial (2021).
Kravchenko, J. E. et al. Small-molecule RETRA suppresses mutant p53-bearing cancer cells through a p73-dependent salvage pathway. Proc. Natl Acad. Sci. USA 105, 6302–6307 (2008).
Zhang, S. et al. Small-molecule NSC59984 restores p53 pathway signaling and antitumor effects against colorectal cancer via p73 activation and degradation of mutant p53. Cancer Res. 75, 3842–3852 (2015).
Xu, J. et al. Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 7, 285–295 (2011).
Zhang, S., Zhou, L. & El-Deiry, W. S. Small-molecule NSC59984 induces mutant p53 degradation through a ROS–ERK2–MDM2 axis in cancer cells. Mol. Cancer Res. 20, 622–636 (2022).
Wang, G. & Fersht, A. R. Propagation of aggregated p53: cross-reaction and coaggregation vs. seeding. Proc. Natl Acad. Sci. USA 112, 2443 (2015).
Soragni, A. et al. A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29, 90–103 (2016).
Palanikumar, L. et al. Protein mimetic amyloid inhibitor potently abrogates cancer-associated mutant p53 aggregation and restores tumor suppressor function. Nat. Commun. 12, 3962 (2021).
Joerger, A. C., Ang, H. C. & Fersht, A. R. Structural basis for understanding oncogenic p53 mutations and designing rescue drugs. Proc. Natl Acad. Sci. USA 103, 15056–15061 (2006).
Boeckler, F. M. et al. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc. Natl Acad. Sci. USA 105, 10360–10365 (2008).
ASCO. https://meetings.asco.org/abstracts-presentations/207594 (2022).
Yu, X., Vazquez, A., Levine, A. J. & Carpizo, D. R. Allele-specific p53 mutant reactivation. Cancer Cell 21, 614–625 (2012).
Yu, X. et al. Thiosemicarbazones functioning as zinc metallochaperones to reactivate mutant p53. Mol. Pharmacol. 91, 567–575 (2017).
Salim, K. Y., Maleki Vareki, S., Danter, W. R. & Koropatnick, J. COTI-2, a novel small molecule that is active against multiple human cancer cell lines in vitro and in vivo. Oncotarget 7, 41363–41379 (2016).
Lindemann, A. et al. COTI-2, a novel thiosemicarbazone derivative, exhibits antitumor activity in HNSCC through p53-dependent and -independent mechanisms. Clin. Cancer Res. 25, 5650–5662 (2019).
Westin, S. N. et al. Abstract CT033: Safety and early efficacy signals for COTI-2, an orally available small molecule targeting p53, in a phase I trial of recurrent gynecologic cancer. Cancer Res. 78, CT033 (2018).
Chen, S. et al. Arsenic trioxide rescues structural p53 mutations through a cryptic allosteric site. Cancer Cell 39, 225–239.e8 (2021). This paper reports that ATO, used for treating acute promyelocytic leukaemia, can reactivate structural p53 mutants and induce p53-dependent tumour suppression.
Oliner, J. D., Kinzler, K. W., Meltzer, P. S., George, D. L. & Vogelstein, B. Amplification of a gene encoding a p53-associated protein in human sarcomas. Nature 358, 80–83 (1992).
Oliner, J. D., Saiki, A. Y. & Caenepeel, S. The role of MDM2 amplification and overexpression in tumorigenesis. Cold Spring Harb. Perspect. Med. 6, a026336 (2016).
Vassilev, L. T. et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848 (2004).
Vu, B. et al. Discovery of RG7112: a small-molecule MDM2 inhibitor in clinical development. ACS Med. Chem. Lett. 4, 466–469 (2013).
Ray-Coquard, I. et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: an exploratory proof-of-mechanism study. Lancet Oncol. 13, 1133–1140 (2012).
Andreeff, M. et al. Results of the phase 1 trial of RG7112, a small-molecule MDM2 antagonist in leukemia. Clin. Cancer Res. 22, 868 (2016).
Kim, E. S. & Shohet, J. M. Reactivation of p53 via MDM2 inhibition. Cell Death Dis. 6, e1936 (2015).
Iancu-Rubin, C. et al. Activation of p53 by the MDM2 inhibitor RG7112 impairs thrombopoiesis. Exp. Hematol. 42, 137–145.e5 (2014).
Xue, Y. et al. Bortezomib stabilizes and activates p53 in proliferative compartments of both normal and tumor tissues in vivo. Cancer Res. 79, 3595–3607 (2019).
Yee, K. et al. Murine double minute 2 inhibition alone or with cytarabine in acute myeloid leukemia: results from an idasanutlin phase 1/1b study. Leuk. Res. 100, 106489 (2021).
Montesinos, P. et al. MIRROS: a randomized, placebo-controlled, phase III trial of cytarabine ± idasanutlin in relapsed or refractory acute myeloid leukemia. Future Oncol. 16, 807–815 (2020).
Konopleva, M. Y. et al. Idasanutlin plus cytarabine in relapsed or refractory acute myeloid leukemia: results of the MIRROS trial. Blood Adv. 6, 4147–4156 (2022).
Mascarenhas, J. et al. Oral idasanutlin in patients with polycythemia vera. Blood 134, 525–533 (2019).
Mascarenhas, J. et al. Safety and efficacy of idasanutlin in patients (pts) with hydroxyurea (HU)-resistant/intolerant polycythemia vera (PV): results of an international phase II study. Blood 136, 29–31 (2020).
Fang, D. D. et al. MDM2 inhibitor APG-115 exerts potent antitumor activity and synergizes with standard-of-care agents in preclinical acute myeloid leukemia models. Cell Death Disco. 7, 90 (2021).
Yi, H. et al. A novel small molecule inhibitor of MDM2-p53 (APG-115) enhances radiosensitivity of gastric adenocarcinoma. J. Exp. Clin. Cancer Res. 37, 97 (2018).
Sun, D. et al. Discovery of AMG 232, a potent, selective, and orally bioavailable MDM2-p53 inhibitor in clinical development. J. Med. Chem. 57, 1454–1472 (2014).
Canon, J. et al. The MDM2 inhibitor AMG 232 demonstrates robust antitumor efficacy and potentiates the activity of p53-inducing cytotoxic agents. Mol. Cancer Ther. 14, 649–658 (2015).
Verstovsek, S. et al. BOREAS: a global phase 3 study of KRT-232, a first-in-class murine double minute 2 (MDM2) inhibitor in TP53WT relapsed/refractory (R/R) myelofibrosis (MF). J. Clin. Oncol. 39 (suppl.15), TPS7057 (2021).
Erba, H. P. et al. Phase 1b study of the MDM2 inhibitor AMG 232 with or without trametinib in relapsed/refractory acute myeloid leukemia. Blood Adv. 3, 1939–1949 (2019).
Jeay, S. et al. Dose and schedule determine distinct molecular mechanisms underlying the efficacy of the p53-MDM2 inhibitor HDM201. Cancer Res. 78, 6257–6267 (2018).
Gounder, M. M. et al. A phase 1 study of the MDM2 inhibitor DS-3032b in patients (pts) with advanced solid tumors and lymphomas. J. Clin. Oncol. 34 (suppl.15), 2581 (2016).
Wade, M., Li, Y. C. & Wahl, G. M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13, 83–96 (2013).
Carvajal, L. A. et al. Dual inhibition of MDMX and MDM2 as a therapeutic strategy in leukemia. Sci. Transl. Med. 10, 3003 (2018).
Tan, Y. S., Lane, D. P. & Verma, C. S. Stapled peptide design: principles and roles of computation. Drug. Discov. Today 21, 1642–1653 (2016).
Bernal, F. et al. A stapled p53 helix overcomes HDMX-mediated suppression of p53. Cancer Cell 18, 411–422 (2010).
Li, Y. C. et al. A versatile platform to analyze low-affinity and transient protein-protein interactions in living cells in real time. Cell Rep. 9, 1946–1958 (2014).
Chang, Y. S. et al. Stapled α-helical peptide drug development: a potent dual inhibitor of MDM2 and MDMX for p53-dependent cancer therapy. Proc. Natl Acad. Sci. USA 110, E3445–E3454 (2013).
Pairawan, S. et al. First in class dual MDM2/MDMX inhibitor ALRN-6924 enhances antitumor efficacy of chemotherapy in TP53 wild-type hormone receptor-positive breast cancer models. Breast Cancer Res 23, 29 (2021).
Meric-Bernstam, F. et al. Phase I trial of a novel stapled peptide ALRN-6924 disrupting MDMX- and MDM2-mediated inhibition of WT p53 in patients with solid tumors and lymphomas. J. Clin. Oncol. 35, 2505 (2017).
Cheng, X. et al. Leveraging the multivalent p53 peptide-MdmX interaction to guide the improvement of small molecule inhibitors. Nat. Commun. 13, 1087 (2022).
Mares, A. et al. Extended pharmacodynamic responses observed upon PROTAC-mediated degradation of RIPK2. Commun. Biol. 3, 140 (2020).
Schneekloth, A. R., Pucheault, M., Tae, H. S. & Crews, C. M. Targeted intracellular protein degradation induced by a small molecule: En route to chemical proteomics. Bioorg. Med. Chem. Lett. 18, 5904–5908 (2008).
Hines, J., Lartigue, S., Dong, H., Qian, Y. & Crews, C. M. MDM2-recruiting PROTAC offers superior, synergistic antiproliferative activity via simultaneous degradation of BRD4 and stabilization of p53. Cancer Res. 79, 251–262 (2019).
Li, Y. et al. Discovery of MD-224 as a first-in-class, highly potent, and efficacious proteolysis targeting chimera murine double minute 2 degrader capable of achieving complete and durable tumor regression. J. Med. Chem. 62, 448–466 (2019).
Wang, B. et al. Development of MDM2 degraders based on ligands derived from Ugi reactions: lessons and discoveries. Eur. J. Med. Chem. 219, 113425 (2021).
He, S. et al. Homo-PROTAC mediated suicide of MDM2 to treat non-small cell lung cancer. Acta Pharm. Sin. B 11, 1617–1628 (2021).
Issaeva, N. et al. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat. Med. 10, 1321–1328 (2004).
Zhao, C. Y., Szekely, L., Bao, W. & Selivanova, G. Rescue of p53 function by small-molecule RITA in cervical carcinoma by blocking E6-mediated degradation. Cancer Res. 70, 3372–3381 (2010).
Weilbacher, A., Gutekunst, M., Oren, M., Aulitzky, W. E. & Van Der Kuip, H. RITA can induce cell death in p53-defective cells independently of p53 function via activation of JNK/SAPK and p38. Cell Death Dis. 5, e1318 (2014).
Wiegering, A. et al. Reactivating p53 and inducing tumor apoptosis (RITA) enhances the response of RITA-sensitive colorectal cancer cells to chemotherapeutic agents 5-fluorouracil and oxaliplatin. Neoplasia 19, 301–309 (2017).
Yang, J. et al. Small-molecule activation of p53 blocks hypoxia-inducible factor 1alpha and vascular endothelial growth factor expression in vivo and leads to tumor cell apoptosis in normoxia and hypoxia. Mol. Cell. Biol. 29, 2243–2253 (2009).
Celegato, M. et al. A novel small-molecule inhibitor of the human papillomavirus E6–p53 interaction that reactivates p53 function and blocks cancer cells growth. Cancer Lett. 470, 115–125 (2020).
Zhang, J. et al. A small-molecule inhibitor of MDMX suppresses cervical cancer cells via the inhibition of E6–E6AP–p53 axis. Pharmacol. Res. 177, 106128 (2022).
Donehower, L. A. et al. Integrated analysis of TP53 gene and pathway alterations in the cancer genome atlas. Cell Rep. 28, 1370–1384.e5 (2019).
Bidou, L., Bugaud, O., Belakhov, V., Baasov, T. & Namy, O. Characterization of new-generation aminoglycoside promoting premature termination codon readthrough in cancer cells. RNA Biol. 14, 378–388 (2017).
Floquet, C., Deforges, J., Rousset, J. P. & Bidou, L. Rescue of non-sense mutated p53 tumor suppressor gene by aminoglycosides. Nucleic Acids Res. 39, 3350–3362 (2011).
Martin, L. et al. Identification and characterization of small molecules that inhibit nonsense-mediated rna decay and suppress nonsense p53 mutations. Cancer Res. 74, 3104–3113 (2014).
Dabrowski, M., Bukowy-Bieryllo, Z. & Zietkiewicz, E. Advances in therapeutic use of a drug-stimulated translational readthrough of premature termination codons. Mol. Med. 24, 25 (2018).
Alexandrova, E. M. et al. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 523, 352–356 (2015).
Padmanabhan, A. et al. USP15-dependent lysosomal pathway controls p53-R175H turnover in ovarian cancer cells. Nat. Commun. 9, 1270 (2018).
Hassin, O. et al. Different hotspot p53 mutants exert distinct phenotypes and predict outcome of colorectal cancer patients. Nat. Commun. 13, 2800 (2022).
Mukherjee, S. et al. Cross-talk between mutant p53 and p62/SQSTM1 augments cancer cell migration by promoting the degradation of cell adhesion proteins. Proc. Natl Acad. Sci. USA 119, e2119644119 (2022).
Kennedy, M. C. & Lowe, S. W. Mutant p53: it’s not all one and the same. Cell Death Differ. 29, 983–987 (2022).
Alvarado-Ortiz, E. et al. Mutant p53 gain-of-function: role in cancer development, progression, and therapeutic approaches. Front. Cell Dev. Biol. 8, 607670 (2021).
Lauwen, M. M. et al. Self-tolerance does not restrict the CD4+ T-helper response against the p53 tumor antigen. Cancer Res. 68, 893–900 (2008).
Chikamatsu, K. et al. Generation of anti-p53 cytotoxic T lymphocytes from human peripheral blood using autologous dendritic cells. Clin. Cancer Res. 5, 1281–1288 (1999).
Röpke, M. et al. Spontaneous human squamous cell carcinomas are killed by a human cytotoxic T lymphocyte clone recognizing a wild-type p53-derived peptide. Proc. Natl Acad. Sci. USA 93, 14704–14707 (1996).
Vierboom, M. P. M. et al. Tumor eradication by wild-type p53-specific cytotoxic T lymphocytes. J. Exp. Med. 186, 695–704 (1997).
Speetjens, F. M. et al. Induction of p53-specific immunity by a p53 synthetic long peptide vaccine in patients treated for metastatic colorectal cancer. Clin. Cancer Res. 15, 1086–1095 (2009).
Vermeij, R. et al. Potentiation of a p53-SLP vaccine by cyclophosphamide in ovarian cancer: a single-arm phase II study. Int. J. Cancer 131, E670–E680 (2012).
Leffers, N. et al. Long-term clinical and immunological effects of p53-SLP® vaccine in patients with ovarian cancer. Int. J. Cancer 130, 105–112 (2012).
Hardwick, N. R. et al. p53MVA therapy in patients with refractory gastrointestinal malignancies elevates p53-specific CD8+ T-cell responses. Clin. Cancer Res. 20, 4459–4470 (2014).
Hardwick, N. R. et al. p53-Reactive T cells are associated with clinical benefit in patients with platinum-resistant epithelial ovarian cancer after treatment with a p53 vaccine and gemcitabine chemotherapy. Clin. Cancer Res. 24, 1315–1325 (2018).
Barfoed, A. M. et al. Cytotoxic T-lymphocyte clones, established by stimulation with the HLA-A2 binding p5365-73 wild type peptide loaded on dendritic cells In vitro, specifically recognize and lyse HLA-A2 tumour cells overexpressing the p53 protein. Scand. J. Immunol. 51, 128–133 (2000).
Antonia, S. J. et al. Combination of p53 cancer vaccine with chemotherapy in patients with extensive stage small cell lung cancer. Clin. Cancer Res. 12, 878–887 (2006).
Chiappori, A. A., Soliman, H., Janssen, W. E., Antonia, S. J. & Gabrilovich, D. I. INGN-225: a dendritic cell-based p53 vaccine (Ad.p53-DC) in small cell lung cancer: observed association between immune response and enhanced chemotherapy effect. Expert. Opin. Biol. Ther. 10, 983–991 (2010).
Chiappori, A. A. et al. Randomized-controlled phase II trial of salvage chemotherapy after immunization with a TP53-transfected dendritic cell-based vaccine (Ad.p53-DC) in patients with recurrent small cell lung cancer. Cancer Immunol. Immunother. 68, 517–527 (2019).
Met, Ö., Balslev, E., Flyger, H. & Svane, I. M. High immunogenic potential of p53 mRNA-transfected dendritic cells in patients with primary breast cancer. Breast Cancer Res. Treat. 125, 395–406 (2010).
Li, D. et al. Development of a T-cell receptor mimic antibody against wild-type p53 for cancer immunotherapy. Cancer Res. 77, 2699–2711 (2017).
Low, L., Goh, A., Koh, J., Lim, S. & Wang, C. I. Targeting mutant p53-expressing tumours with a T cell receptor-like antibody specific for a wild-type antigen. Nat. Commun. 10, 5382 (2019).
Cheever, M. A. et al. The prioritization of cancer antigens: a National Cancer Institute pilot project for the acceleration of translational research. Clin. Cancer Res. 15, 5323–5337 (2009).
Balachandran, V. et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516 (2017).
Deniger, D. C. et al. T-cell responses to TP53 ‘Hotspot’ mutations and unique neoantigens expressed by human ovarian cancers. Clin. Cancer Res. 24, 5562–5573 (2018).
Malekzadeh, P. et al. Neoantigen screening identifies broad TP53 mutant immunogenicity in patients with epithelial cancers. J. Clin. Invest. 129, 1109–1114 (2019).
Malekzadeh, P. et al. Antigen experienced T cells from peripheral blood recognize p53 neoantigens. Clin. Cancer Res. 26, 1267–1276 (2020).
You, G. et al. Bispecific antibodies: a smart arsenal for cancer immunotherapies. Vaccines 9, 724 (2021).
Hsiue, E. H. C. et al. Targeting a neoantigen derived from a common TP53 mutation. Science 371, eabc8697 (2021). This paper reports a bispecific antibody, highly specific to the p53(R175H) mutation, which activates T cells to kill cancer cells presenting a p53(R175H)-derived neoantigen in vitro and in vivo.
Levine, A. J. P53 and the immune response: 40 years of exploration-a plan for the future. Int. J. Mol. Sci. 21, 541 (2020).
Cortez, M. A. et al. PDL1 regulation by p53 via miR-34. J. Natl Cancer Inst. 108, djv303 (2015). This paper reports that wtp53 can downregulate PDL1 in cancer cells, suggesting a mechanism whereby loss of wtp53 can render such cells more refractory to immune attack.
Textor, S. et al. Human NK cells are alerted to induction of p53 in cancer cells by upregulation of the NKG2D ligands ULBP1 and ULBP2. Cancer Res. 71, 5998–6009 (2011).
Zhou, X. et al. Pharmacological activation of p53 triggers viral mimicry response thereby abolishing tumor immune evasion and promoting anti-tumor immunity. Cancer Discov. 11, 3090–3105 (2021).
Maddalena, M. et al. TP53 missense mutations in PDAC are associated with enhanced fibrosis and an immunosuppressive microenvironment. Proc. Natl Acad. Sci. USA 118, e2025631118 (2021).
PMVPharma. PMV Pharmaceuticals Announces a Clinical Trial Collaboration with Merck to Evaluate PC14586 in Combination with KEYTRUDA® (pembrolizumab) in Patients with Advanced Solid Tumors. GlobeNewswire (18 July 2022); go.nature.com/3fyQKDo
Moskovits, N., Kalinkovich, A., Bar, J., Lapidot, T. & Oren, M. p53 Attenuates cancer cell migration and invasion through repression of SDF-1/CXCL12 expression in stromal fibroblasts. Cancer Res. 66, 10671–10676 (2006).
Fang, D. D. et al. MDM2 inhibitor APG-115 synergizes with PD-1 blockade through enhancing antitumor immunity in the tumor microenvironment. J. Immunother. Cancer 7, 327 (2019). This paper describes the enhanced antitumour effect of combining a p53 activating molecule (APG-115) with an anti-PD1 antibody, attributed to the effect of APG-115 on the tumour immune microenvironment.
Tolcher, A. W. et al. Preliminary results of a phase II study of alrizomadlin (APG-115), a novel, small-molecule MDM2 inhibitor, in combination with pembrolizumab in patients (pts) with unresectable or metastatic melanoma or advanced solid tumors that have failed immuno-oncologic (I-O) drugs. J. Clin. Oncol. 39, 2506 (2021).
Kim, S.-S., Harford, J. B., Moghe, M., Rait, A. & Chang, E. H. Combination with SGT-53 overcomes tumor resistance to a checkpoint inhibitor. Oncoimmunology 7, e1484982 (2018).
Chada, S. et al. Tumor suppressor immune gene therapy to reverse immunotherapy resistance. Cancer Gene Ther. 29, 825–834 (2022).
Wang, D. & Wang, K. An overview of development in gene therapeutics in China. Gene Ther. 27, 338–348 (2020).
Goswami, R. et al. Gene therapy leaves a vicious cycle. Front. Oncol. 9, 297 (2019).
Zhang, W. W. et al. The first approved gene therapy product for cancer ad-p53 (gendicine): 12 years in the clinic. Hum. Gene Ther. 29, 160–179 (2018).
Xia, Y., Du, Z., Wang, X. & Li, X. Treatment of uterine sarcoma with rAd-p53 (gendicine) followed by chemotherapy: clinical study of TP53 gene therapy. Hum. Gene Ther. 29, 242–250 (2018).
Li, Y. et al. Selective intra-arterial infusion of rAd-p53 with chemotherapy for advanced oral cancer: a randomized clinical trial. BMC Med. 12, 16 (2014).
Liu, S. et al. Randomized, controlled phase II study of post-surgery radiotherapy combined with recombinant adenoviral human p53 gene therapy in treatment of oral cancer. Cancer Gene Ther. 20, 375–378 (2013).
Gabrilovich, D. I. INGN 201 (Advexin): adenoviral p53 gene therapy for cancer. Expert Opin. Biol. Ther. 6, 823–832 (2006).
Atencio, I. A. et al. Biological activities of a recombinant adenovirus p53 (SCH 58500) administered by hepatic arterial infusion in a phase 1 colorectal cancer trial. Cancer Gene Ther. 13, 169–181 (2006).
Buller, R. E. et al. A phase I/II trial of rAd/p53 (SCH 58500) gene replacement in recurrent ovarian cancer. Cancer Gene Ther. 9, 553–566 (2002).
Zeimet, A. G. & Marth, C. Why did p53 gene therapy fail in ovarian cancer? Lancet Oncol. 4, 415–422 (2003).
Li, Y. et al. Expert consensus on the clinical application of recombinant adenovirus human p53 for head and neck cancers. Int. J. Oral. Sci. 13, 38 (2021).
Tamura, R. E. et al. Induction of oxidants distinguishes susceptibility of prostate carcinoma cell lines to p53 gene transfer mediated by an improved adenoviral vector. Hum. Gene Ther. 28, 639–653 (2017).
Chen, S. et al. A surface charge-switchable and folate modified system for co-delivery of proapoptosis peptide and p53 plasmid in cancer therapy. Biomaterials 77, 149–163 (2016).
Misra, S. K., Naz, S., Kondaiah, P. & Bhattacharya, S. A cationic cholesterol based nanocarrier for the delivery of p53-EGFP-C3 plasmid to cancer cells. Biomaterials 35, 1334–1346 (2014).
Rejeeth, C. & Kannan, S. p53 Gene therapy of human breast carcinoma: using a transferrin-modified silica nanoparticles. Breast Cancer 23, 101–110 (2016).
Kim, S. S., Rait, A., Kim, E., Pirollo, K. F. & Chang, E. H. A tumor-targeting p53 nanodelivery system limits chemoresistance to temozolomide prolonging survival in a mouse model of glioblastoma multiforme. Nanomedicine 11, 301–311 (2015).
Senzer, N. et al. Phase I study of a systemically delivered p53 nanoparticle in advanced solid tumors. Mol. Ther. 21, 1096–1103 (2013).
Pirollo, K. F. et al. Safety and efficacy in advanced solid tumors of a targeted nanocomplex carrying the p53 gene used in combination with docetaxel: a phase 1b study. Mol. Ther. 24, 1697–1706 (2016).
Kong, N. et al. Synthetic mRNA nanoparticle-mediated restoration of p53 tumor suppressor sensitizes p53-deficient cancers to mTOR inhibition. Sci. Transl. Med. 11, 1565 (2019). This paper describes the use of synthetic p53 mRNA as a potential anticancer treatment.
Xiao, Y. et al. Combining p53 mRNA nanotherapy with immune checkpoint blockade reprograms the immune microenvironment for effective cancer therapy. Nat. Commun. 13, 758 (2022).
Sobol, R. E. et al. Meta-analysis of adenoviral p53 gene therapy clinical trials in recurrent head and neck squamous cell carcinoma. medRxiv https://doi.org/10.1101/2021.01.06.20248743 (2021).
Martinez, L. A. et al. Synthetic small inhibiting RNAs: efficient tools to inactivate oncogenic mutations and restore p53 pathways. Proc. Natl Acad. Sci. USA 99, 14849–14854 (2002).
Ubby, I. et al. Cancer therapeutic targeting using mutant–p53-specific siRNAs. Oncogene 38, 3415–3427 (2019).
Mirgayazova, R. et al. Therapeutic editing of the TP53 gene: is CRISPR/Cas9 an option? Genes 11, 704 (2020).
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A. & Liu, D. R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424 (2016).
Haapaniemi, E., Botla, S., Persson, J., Schmierer, B. & Taipale, J. CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 24, 927–930 (2018).
Ihry, R. J. et al. p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat. Med. 24, 939–946 (2018).
Enache, O. M. et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 52, 662–668 (2020).
Michaelis, M. et al. Adaptation of cancer cells from different entities to the MDM2 inhibitor nutlin-3 results in the emergence of p53-mutated multi-drug-resistant cancer cells. Cell Death Dis. 2, e243 (2011).
Chapeau, E. A. et al. Resistance mechanisms to TP53-MDM2 inhibition identified by in vivo piggyBac transposon mutagenesis screen in an Arf−/− mouse model. Proc. Natl Acad. Sci. USA 114, 3151–3156 (2017).
Latif, A. L. et al. BRD4-mediated repression of p53 is a target for combination therapy in AML. Nat. Commun. 12, 241 (2021).
Pan, R. et al. Synthetic lethality of combined Bcl-2 inhibition and p53 activation in AML: mechanisms and superior antileukemic efficacy. Cancer Cell 32, 748–760.e6 (2017).
Daver, N. G. et al. Safety, efficacy, pharmacokinetic (PK) and biomarker analyses of BCL2 inhibitor venetoclax (Ven) plus MDM2 inhibitor idasanutlin (idasa) in patients (pts) with relapsed or refractory (R/R) AML: a phase Ib, non-randomized, open-label study. Blood 132, 767–767 (2018).
Fischer, M. Mice are not humans: the case of p53. Trends Cancer 7, 12–14 (2021).
Horejs, C. Organ chips, organoids and the animal testing conundrum. Nat. Rev. Mater. 6, 372–373 (2021).
Gavert, N. et al. Ex vivo organotypic cultures for synergistic therapy prioritization identify patient-specific responses to combined MEK and Src inhibition in colorectal cancer. Nat. Cancer 3, 219–231 (2022).
Hong, D. S. et al. KRAS G12C inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 383, 1207–1217 (2020).
Degtjarik, O. et al. Structural basis of reactivation of oncogenic p53 mutants by a small molecule: methylene quinuclidinone (MQ). Nat. Commun. 12, 7057 (2021).
Acknowledgements
Work in the authors’ lab is supported in part by grants from the Dr Miriam and Sheldon G. Adelson Medical Research Foundation, a Center of Excellence from the Israel Science Foundation, the Israel Science Foundation within the Israel Precision Medicine Partnership programme, the Rising Tide Foundation, Quintrigen, The Robert Bosch Stiftung GmbH and the Berthold Leibinger Stiftung GmbH, the United States–Israel Binational Science Foundation (BSF), Jerusalem, Israel, and the Moross Integrated Cancer Center.
Author information
Authors and Affiliations
Contributions
Both authors contributed equally to all aspects of this Review.
Corresponding author
Ethics declarations
Competing interests
M.O. consults for Quintrigen.
Peer review
Peer review information
Nature Reviews Drug Discovery thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Michael acceptor
-
Michael addition is a process of addition of a carbanion or another nucleophile to an α,β-unsaturated carbonyl compound. Michael acceptors are the substituent groups on the activated unsaturated compound. The ability of Michael acceptors to form conjugates with peptides bearing nucleophilic groups provides them with a broad spectrum of potential biological effects.
- Stapled peptides
-
Short peptides locked in a specific conformation by an external brace formed by covalent binding between two amino acid side chains. This allows the molecule to be kept in an α-helical structure with high hydrophobicity, facilitating the crossing of biological membranes.
- Bispecific antibodies
-
Artificial antibodies that possess two binding sites, designed to recognize two different epitopes or antigens. One of the major applications of bispecific antibodies is to bridge two different cell types.
- Indels
-
Insertions or deletions of one or more nucleotides in a DNA sequence.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hassin, O., Oren, M. Drugging p53 in cancer: one protein, many targets. Nat Rev Drug Discov 22, 127–144 (2023). https://doi.org/10.1038/s41573-022-00571-8
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41573-022-00571-8
This article is cited by
-
Biogenic synthesis of gold nanoparticles using Coleus forskohlii leaf extract enhances anticancer activity against HT-29 cells and modulates the p53 regulated immune microenvironment in colon adenocarcinoma
Scientific Reports (2026)
-
The role of cellular senescence-related genes in ischemia–reperfusion injury and the identification of their biomarkers
Scientific Reports (2026)
-
RHBDD1 induces cell cycle arrest in TP53-mutant NSCLC cells by promoting endoplasmic reticulum-associated degradation of p53 and DNA-PKcs
European Journal of Medical Research (2025)
-
Advancements in understanding tumor-resident bacteria and their application in cancer therapy
Military Medical Research (2025)
-
Tendomodulin in pan-cancer analysis: exploring its impact on immune modulation and uncovering functional insights in colorectal cancer
BMC Cancer (2025)