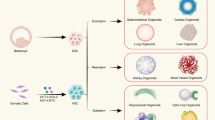
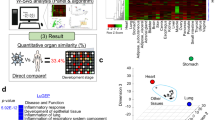
Abstract
Organoids are 3D structures derived from stem cells that recapitulate key architectural and functional aspects of the corresponding tissue. Compared with conventional 2D cell lines, human organoids provide experimental models that more closely reflect human physiology. Their ability to capture the complexity and heterogeneity of human tissues enables the study of disease mechanisms, drug efficacy and toxicity. When generated from patient material, organoids also allow the assessment of individual drug responses. In this Review, we explore the utility of organoids in drug discovery. We outline current methodologies for generating and maintaining organoids, examine their applications in disease modelling, drug screening and safety evaluation, and consider regulatory aspects and the challenges for their broader adoption in drug discovery.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lee, G. Y., Kenny, P. A., Lee, E. H. & Bissell, M. J. Three-dimensional culture models of normal and malignant breast epithelial cells. Nat. Methods 4, 359–365 (2007). This article represents a breakthrough in the 3D culture of epithelial cells using Matrigel as a scaffold.
Martin, G. R. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl Acad. Sci. USA 78, 7634–7638 (1981).
Thomson, J. A. et al. Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147 (1998).
Chambers, S. M. et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 27, 275–280 (2009).
Lian, X. et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc. Natl Acad. Sci. USA 109, E1848–E1857 (2012).
Takahashi, K. & Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 (2006).
Yamanaka, S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell 10, 678–684 (2012).
Kadoshima, T. et al. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell–derived neocortex. Proc. Natl Acad. Sci. USA 110, 20284–20289 (2013).
Eiraku, M. et al. Self-organizing optic-cup morphogenesis in three-dimensional culture. Nature 472, 51–56 (2011). The findings of this study reveal that ESC aggregates can autonomously organize into structured 3D cultures, thereby establishing the first PS cell-derived organoid model.
Nakano, T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012).
Muguruma, K., Nishiyama, A., Kawakami, H., Hashimoto, K. & Sasai, Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 10, 537–550 (2015).
Lancaster, M. A. et al. Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379 (2013).
Cowan, C. S. et al. Cell types of the human retina and its organoids at single-cell resolution. Cell 182, 1623–1640.e1634 (2020).
Miller, A. J. et al. Generation of lung organoids from human pluripotent stem cells in vitro. Nat. Protoc. 14, 518–540 (2019).
Jacob, A. et al. Differentiation of human pluripotent stem cells into functional lung alveolar epithelial cells. Cell Stem Cell 21, 472–488.e410 (2017).
Chen, Y.-W. et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 19, 542–549 (2017).
McCauley, K. B. et al. Efficient derivation of functional human airway epithelium from pluripotent stem cells via temporal regulation of wnt signaling. Cell Stem Cell 20, 844–857.e846 (2017).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Hannan, N. R. F., Segeritz, C.-P., Touboul, T. & Vallier, L. Production of hepatocyte-like cells from human pluripotent stem cells. Nat. Protoc. 8, 430–437 (2013).
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2010).
Till, J. E. & McCulloch, E. A. A direct measurement of the radiation sensitivity of normal mouse bone marrow cells1. Radiat. Res. 175, 145–149 (2011).
Toma, J. G. et al. Isolation of multipotent adult stem cells from the dermis of mammalian skin. Nat. Cell Biol. 3, 778–784 (2001).
Schermer, A., Galvin, S. & Sun, T. T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J. Cell Biol. 103, 49–62 (1986).
Barker, N. et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature 449, 1003–1007 (2007).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). This proof-of-concept study reports the first intestinal organoids derived from TSCs, demonstrating the potential use of TSCs to generate organoids.
Kim, J., Koo, B.-K. & Knoblich, J. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
Sugimoto, S. et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature 592, 99–104 (2021).
Doke, S. K. & Dhawale, S. C. Alternatives to animal testing: a review. Saudi Pharm. J. 23, 223–229 (2015).
Stresser, D. M. et al. Towards in vitro models for reducing or replacing the use of animals in drug testing. Nat. Biomed. Eng. 8, 930–935 (2023).
Paul, R., S.5002, FDA Modernization Act 2.0. Congress.gov https://www.congress.gov/bill/117th-congress/senate-bill/5002 (2022).
Carter, E. L. B., H.R.7248, FDA Modernization Act 3.0. Congress.gov https://www.congress.gov/bill/118th-congress/house-bill/7248/text (2023).
US Food and Drug Administration. FDA announces plan to phase out animal testing requirement for monoclonal antibodies and other drugs. fda.gov https://www.fda.gov/news-events/press-announcements/fda-announces-plan-phase-out-animal-testing-requirement-monoclonal-antibodies-and-other-drugs (2025).
Tsui, L. C. et al. Cystic fibrosis locus defined by a genetically linked polymorphic DNA marker. Science 230, 1054–1057 (1985).
Collins, F. S. Cystic fibrosis: molecular biology and therapeutic implications. Science 256, 774–779 (1992).
Greger, R. Role of CFTR in the colon. Annu. Rev. Physiol. 62, 467–491 (2000).
Lopes-Pacheco, M. CFTR modulators: the changing face of cystic fibrosis in the era of precision medicine. Front. Pharmacol. 10, 1662 (2020).
Guo, J., Garratt, A. & Hill, A. Worldwide rates of diagnosis and effective treatment for cystic fibrosis. J. Cyst. Fibros. 21, 456–462 (2022).
Wainwright, C. E. et al. Lumacaftor–ivacaftor in patients with cystic fibrosis homozygous for Phe508delCFTR. N. Engl. J. Med. 373, 220–231 (2015).
Allen, L. et al. Future therapies for cystic fibrosis. Nat. Commun. 14, 693 (2023).
Ramalho, A. S., Amato, F. & Gentzsch, M. Patient-derived cell models for personalized medicine approaches in cystic fibrosis. J. Cyst. Fibros. 22, S32–S38 (2023).
Lavelle, G. M., White, M. M., Browne, N., McElvaney, N. G. & Reeves, E. P. Animal models of cystic fibrosis pathology: phenotypic parallels and divergences. Biomed. Res. Int. 2016, 5258727 (2016).
Dekkers, J. F. et al. A functional CFTR assay using primary cystic fibrosis intestinal organoids. Nat. Med. 19, 939–945 (2013). This study develops the first in vitro assay to involve organoids to assess CFTR functionality, which has been further applied in personalized CF treatment.
Sachs, N. et al. Long-term expanding human airway organoids for disease modeling. EMBO J. 38, e100300 (2019).
Geurts, M. H. et al. CRISPR-based adenine editors correct nonsense mutations in a cystic fibrosis organoid biobank. Cell Stem Cell 26, 503–510.e507 (2020). This research on PDOs highlights the potential of CRISPR-based gene editing technologies to correct pathogenic mutations associated with CF.
Furstova, E. et al. Response to elexacaftor/tezacaftor/ivacaftor in intestinal organoids derived from people with cystic fibrosis. J. Cyst. Fibros. 21, 243–245 (2022).
Lefferts, J. W. et al. CFTR function restoration upon elexacaftor/tezacaftor/ivacaftor treatment in patient-derived intestinal organoids with rare CFTR genotypes. Int. J. Mol. Sci. 24, 14539 (2023).
Berkers, G. et al. Rectal organoids enable personalized treatment of cystic fibrosis. Cell Rep. 26, 1701–1708.e1703 (2019).
van der Vaart, J. et al. Modelling of primary ciliary dyskinesia using patient-derived airway organoids. EMBO Rep. 22, e52058 (2021).
Lim, K. et al. Organoid modeling of human fetal lung alveolar development reveals mechanisms of cell fate patterning and neonatal respiratory disease. Cell Stem Cell 30, 20–37.e29 (2023).
Leibel, S. L. et al. Reversal of surfactant protein B deficiency in patient specific human induced pluripotent stem cell derived lung organoids by gene therapy. Sci. Rep. 9, 13450 (2019).
Tran, T. et al. A scalable organoid model of human autosomal dominant polycystic kidney disease for disease mechanism and drug discovery. Cell Stem Cell 29, 1083–1101.e1087 (2022).
Xu, Y. et al. Adult human kidney organoids originate from CD24+ cells and represent an advanced model for adult polycystic kidney disease. Nat. Genet. 54, 1690–1701 (2022).
Hiratsuka, K. et al. Organoid-on-a-chip model of human ARPKD reveals mechanosensing pathomechanisms for drug discovery. Sci. Adv. 8, eabq0866 (2022).
Janciauskiene, S. M. et al. The discovery of α1-antitrypsin and its role in health and disease. Respir. Med. 105, 1129–1139 (2011).
Hatipoğlu, U. & Stoller, J. K. α1-Antitrypsin deficiency. Clin. Chest Med. 37, 487–504 (2016).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Gómez-Mariano, G. et al. Liver organoids reproduce alpha-1 antitrypsin deficiency-related liver disease. Hepatol. Int. 14, 127–137 (2019).
Lehmann, V. et al. The potential and limitations of intrahepatic cholangiocyte organoids to study inborn errors of metabolism. J. Inherit. Metab. Dis. 45, 353–365 (2021).
Reed, E., Lutsenko, S. & Bandmann, O. Animal models of Wilson disease. J. Neurochem. 146, 356–373 (2018).
Amarachintha, S. P. et al. Biliary organoids uncover delayed epithelial development and barrier function in biliary atresia. Hepatology 75, 89–103 (2021).
Babu, R. O. et al. Beta-amyloid deposition around hepatic bile ducts is a novel pathobiological and diagnostic feature of biliary atresia. J. Hepatol. 73, 1391–1403 (2020).
Wulansari, N. et al. Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci. Adv. 7, eabb1540 (2021).
Kim, H. et al. Modeling G2019S-LRRK2 sporadic Parkinson’s disease in 3D midbrain organoids. Stem Cell Rep. 12, 518–531 (2019).
Zhao, J. et al. APOE4 exacerbates synapse loss and neurodegeneration in Alzheimer’s disease patient iPSC-derived cerebral organoids. Nat. Commun. 11, 5540 (2020).
Park, J. et al. A 3D human triculture system modeling neurodegeneration and neuroinflammation in Alzheimer’s disease. Nat. Neurosci. 21, 941–951 (2018).
Lin, Y. T. et al. APOE4 causes widespread molecular and cellular alterations associated with Alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 98, 1141–1154.e1147 (2018).
Raja, W. K. et al. Self-organizing 3D human neural tissue derived from induced pluripotent stem cells recapitulate Alzheimer’s disease phenotypes. PLoS ONE 11, e0161969 (2016).
Nakamura, M. et al. Pathological progression induced by the frontotemporal dementia-associated R406W Tau mutation in patient-derived iPSCs. Stem Cell Rep. 13, 684–699 (2019).
Bowles, K. R. et al. ELAVL4, splicing, and glutamatergic dysfunction precede neuron loss in MAPT mutation cerebral organoids. Cell 184, 4547–4563.e4517 (2021).
Shimada, H. et al. A next-generation iPSC-derived forebrain organoid model of tauopathy with tau fibrils by AAV-mediated gene transfer. Cell Rep. Methods 2, 100289 (2022).
Szebenyi, K. et al. Human ALS/FTD brain organoid slice cultures display distinct early astrocyte and targetable neuronal pathology. Nat. Neurosci. 24, 1542–1554 (2021).
Lai, J. D. et al. KCNJ2 inhibition mitigates mechanical injury in a human brain organoid model of traumatic brain injury. Cell Stem Cell 31, 519–536.e518 (2024).
Kang, Y. et al. A human forebrain organoid model of fragile X syndrome exhibits altered neurogenesis and highlights new treatment strategies. Nat. Neurosci. 24, 1377–1391 (2021).
Bear, M. F., Huber, K. M. & Warren, S. T. The mGluR theory of fragile X mental retardation. Trends Neurosci. 27, 370–377 (2004).
Sharma, S. D. et al. Astrocytes mediate cell non-autonomous correction of aberrant firing in human FXS neurons. Cell Rep. 42, 112344 (2023).
Zou, Z. et al. FMRP phosphorylation modulates neuronal translation through YTHDF1. Mol. Cell 83, 4304–4317.e4308 (2023).
Younossi, Z. M. et al. Global epidemiology of nonalcoholic fatty liver disease—Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016).
Loomba, R., Friedman, S. L. & Shulman, G. I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021).
Igarashi, R. et al. Generation of human adult hepatocyte organoids with metabolic functions. Nature https://doi.org/10.1038/s41586-025-08861-y (2025). This study reports the establishment of expandable and functional hepatocyte organoids directly from adult liver tissues.
McCarron, S. et al. Functional characterization of organoids derived from irreversibly damaged liver of patients with NASH. Hepatology 74, 1825–1844 (2021).
Wang, L. et al. Recapitulating lipid accumulation and related metabolic dysregulation in human liver-derived organoids. J. Mol. Med. 100, 471–484 (2022).
Mun, S. J. et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 71, 970–985 (2019).
Ouchi, R. et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30, 374–384.e376 (2019).
Morais, J. B. S. et al. Effect of magnesium supplementation on insulin resistance in humans: a systematic review. Nutrition 38, 54–60 (2017).
Romualdo, G. R. et al. Sorafenib reduces steatosis-induced fibrogenesis in a human 3D co-culture model of non-alcoholic fatty liver disease. Environ. Toxicol. 36, 168–176 (2020).
Kumar, M. et al. A fully defined matrix to support a pluripotent stem cell derived multi-cell-liver steatohepatitis and fibrosis model. Biomaterials 276, 121006 (2021).
Cayo, M. A. et al. A drug screen using human iPSC-derived hepatocyte-like cells reveals cardiac glycosides as a potential treatment for hypercholesterolemia. Cell Stem Cell 20, 478–489.e475 (2017).
Jing, R. et al. A screen using iPSC-derived hepatocytes reveals NAD+ as a potential treatment for mtDNA depletion syndrome. Cell Rep. 25, 1469–1484.e1465 (2018).
Rezvani, M., Vallier, L. & Guillot, A. Modeling nonalcoholic fatty liver disease in the dish using human-specific platforms: strategies and limitations. Cell. Mol. Gastroenterol. Hepatol. 15, 1135–1145 (2023).
Hendriks, D. et al. Engineered human hepatocyte organoids enable CRISPR-based target discovery and drug screening for steatosis. Nat. Biotechnol. 41, 1567–1581 (2023). This representative study integrates the hepatocyte organoid system with phenotypic imaging techniques for enhanced conventional drug screening and CRISPR-based mechanistic exploration.
Ananthakrishnan, A. N. Epidemiology and risk factors for IBD. Nat. Rev. Gastroenterol. Hepatol. 12, 205–217 (2015).
de Lange, K. M. et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat. Genet. 49, 256–261 (2017).
d’Aldebert, E. et al. Characterization of human colon organoids from inflammatory bowel disease patients. Front. Cell Dev. Biol. 8, 363 (2020).
Pavlidis, P. et al. Cytokine responsive networks in human colonic epithelial organoids unveil a molecular classification of inflammatory bowel disease. Cell Rep. 40, 111439 (2022).
Pavlidis, P. et al. Interleukin-22 regulates neutrophil recruitment in ulcerative colitis and is associated with resistance to ustekinumab therapy. Nat. Commun. 13, 5820 (2022).
Niklinska-Schirtz, B. J. et al. Ileal derived organoids from Crohn’s disease patients show unique transcriptomic and secretomic signatures. Cell. Mol. Gastroenterol. Hepatol. 12, 1267–1280 (2021).
Howell, K. J. et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology 154, 585–598 (2018).
Hammoudi, N. et al. Autologous organoid co-culture model reveals T cell-driven epithelial cell death in Crohn’s disease. Front. Immunol. 13, 1008456 (2022).
Iversen, R. & Sollid, L. M. The immunobiology and pathogenesis of celiac disease. Annu. Rev. Pathol. 24, 47–70 (2023).
de Kauwe, A. L. et al. Resistance to celiac disease in humanized HLA-DR3-DQ2-transgenic mice expressing specific anti-gliadin CD4+ T cells. J. Immunol. 182, 7440–7450 (2009).
Abadie, V. et al. IL-15, gluten and HLA-DQ8 drive tissue destruction in coeliac disease. Nature 578, 600–604 (2020).
Santos, A. J. M. et al. A human autoimmune organoid model reveals IL-7 function in coeliac disease. Nature 632, 401–410 (2024). This study introduces the first organoid model for CeD, integrating epithelial and immune cells in a co-culture system to facilitate the identification of potential therapeutic target IL-7.
Casto, C. et al. Hashimoto’s thyroiditis and Graves’ disease in genetic syndromes in pediatric age. Genes 12, 222 (2021).
van der Vaart, J. et al. Adult mouse and human organoids derived from thyroid follicular cells and modeling of Graves’ hyperthyroidism. Proc. Natl Acad. Sci. USA 118, e2117017118 (2021).
Xiao, H. et al. Proteomics and organoid culture reveal the underlying pathogenesis of Hashimoto’s thyroiditis. Front. Immunol. 12, 784975 (2021).
Chan, L. L. Y. et al. The establishment of COPD organoids to study host–pathogen interaction reveals enhanced viral fitness of SARS-CoV-2 in bronchi. Nat. Commun. 13, 7635 (2022).
Wu, X. et al. A transcriptomics-guided drug target discovery strategy identifies receptor ligands for lung regeneration. Sci. Adv. 8, eabj9949 (2022).
Heo, I. et al. Modelling Cryptosporidium infection in human small intestinal and lung organoids. Nat. Microbiol. 3, 814–823 (2018).
Ettayebi, K. et al. Replication of human noroviruses in stem cell-derived human enteroids. Science 353, 1387–1393 (2016). This study highlights the advantage of a human intestinal organoid system in supporting the complete viral life cycle, a capability that remains challenging in traditional 2D cell lines.
Baldridge, M. T., Turula, H. & Wobus, C. E. Norovirus regulation by host and microbe. Trends Mol. Med. 22, 1047–1059 (2016).
Zhang, D. et al. Human intestinal organoids express histo-blood group antigens, bind norovirus VLPs, and support limited norovirus replication. Sci. Rep. 7, 12621 (2017).
Cheetham, S. et al. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J. Virol. 80, 10372–10381 (2006).
Rockx, B. H. G., Bogers, W. M. J. M., Heeney, J. L., van Amerongen, G. & Koopmans, M. P. G. Experimental norovirus infections in non-human primates. J. Med. Virol. 75, 313–320 (2004).
Finkbeiner, S. R. et al. Stem cell-derived human intestinal organoids as an infection model for rotaviruses. mBio 3, e00159–12 (2012).
Saxena, K. et al. Human intestinal enteroids: a new model to study human rotavirus infection, host restriction, and pathophysiology. J. Virol. 90, 43–56 (2016).
Xing, Z. et al. Human gut-on-a-chip supports polarized infection of Coxsackie B1 virus in vitro. PLoS ONE 12, e0169412 (2017).
Drummond, C. G. et al. Enteroviruses infect human enteroids and induce antiviral signaling in a cell lineage-specific manner. Proc. Natl Acad. Sci. USA 114, 1672–1677 (2017).
Tsang, J. O.-L. et al. Development of three-dimensional human intestinal organoids as a physiologically relevant model for characterizing the viral replication kinetics and antiviral susceptibility of enteroviruses. Biomedicines 9, 88 (2021).
Kleber de Oliveira, W. et al. Increase in reported prevalence of microcephaly in infants born to women living in areas with confirmed zika virus transmission during the first trimester of pregnancy - Brazil, 2015. MMWR Morb. Mortal. Wkly Rep. 65, 242–247 (2016).
Garcez, P. P. et al. Zika virus impairs growth in human neurospheres and brain organoids. Science 352, 816–818 (2016).
Qian, X. et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell 165, 1238–1254 (2016).
Cugola, F. R. et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature 534, 267–271 (2016).
Tang, H. et al. Zika virus infects human cortical neural progenitors and attenuates their growth. Cell Stem Cell 18, 587–590 (2016).
Gabriel, E. et al. Recent Zika virus isolates induce premature differentiation of neural progenitors in human brain organoids. Cell Stem Cell 20, 397–406.e395 (2017).
Yoon, K. J. et al. Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell 21, 349–358.e346 (2017).
Liang, Q. et al. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell 19, 663–671 (2016).
Grant, A. et al. Zika virus targets human STAT2 to inhibit type I interferon signaling. Cell Host Microbe 19, 882–890 (2016).
Dang, J. et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell 19, 258–265 (2016).
Zhou, T. et al. High-content screening in hPSC-neural progenitors identifies drug candidates that inhibit Zika virus infection in fetal-like organoids and adult brain. Cell Stem Cell 21, 274–283.e275 (2017).
Xu, M. et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat. Med. 22, 1101–1107 (2016).
Chen, X. et al. In vitro and in vivo inhibition of the host TRPC4 channel attenuates Zika virus infection. EMBO Mol. Med. 16, 1817–1839 (2024).
Milewska, A. et al. Replication of severe acute respiratory syndrome coronavirus 2 in human respiratory epithelium. J. Virol. 94, e00957–20 (2020).
Lamers, M. M. et al. SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54 (2020).
Beumer, J. et al. A CRISPR/Cas9 genetically engineered organoid biobank reveals essential host factors for coronaviruses. Nat. Commun. 12, 5498 (2021).
Altulea, D., Maassen, S., Baranov, M. V., van den Bogaart, G. & Wu, J. What makes (hydroxy)chloroquine ineffective against COVID-19: insights from cell biology. J. Mol. Cell Biol. 13, 175–184 (2021).
Zhou, J. et al. Differentiated human airway organoids to assess infectivity of emerging influenza virus. Proc. Natl Acad. Sci. USA 115, 6822–6827 (2018).
Bui, C. H. T. et al. Risk assessment for highly pathogenic avian influenza A(H5N6/H5N8) clade 2.3.4.4 viruses. Emerg. Infect. Dis. 27, 2619–2627 (2021).
Li, C. et al. Human respiratory organoids sustained reproducible propagation of human rhinovirus C and elucidation of virus-host interaction. Nat. Commun. 15, 10772 (2024).
Sato, T. et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 141, 1762–1772 (2011).
van de Wetering, M. et al. Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945 (2015). This proof-of-concept study describes the establishment of PDTO biobanks for CRCs, highlighting their ability to preserve cellular heterogeneity and genetic diversity, thus proving valuable for drug screening applications.
Roper, J. et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 35, 569–576 (2017).
Matano, M. et al. Modeling colorectal cancer using CRISPR–Cas9–mediated engineering of human intestinal organoids. Nat. Med. 21, 256–262 (2015). As proof of concept, this study reports the generation of CRC organoids from wild-type human organoid models through defined CRISPR gene editing of CRC-associated genes, and further demonstrates pathological similarities between the transplanted CRC organoids and actual tumours.
Seino, T. et al. Human pancreatic tumor organoids reveal loss of stem cell niche factor dependence during disease progression. Cell Stem Cell 22, 454–467.e456 (2018).
Sachs, N. et al. A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e310 (2018).
Yan, H. H. N. et al. A comprehensive human gastric cancer organoid biobank captures tumor subtype heterogeneity and enables therapeutic screening. Cell Stem Cell 23, 882–897.e811 (2018).
Jacob, F. et al. A patient-derived glioblastoma organoid model and Biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 180, 188–204.e122 (2020).
Driehuis, E. et al. Pancreatic cancer organoids recapitulate disease and allow personalized drug screening. Proc. Natl Acad. Sci. USA 116, 26580–26590 (2019).
Vlachogiannis, G. et al. Patient-derived organoids model treatment response of metastatic gastrointestinal cancers. Science 359, 920–926 (2018).
Yao, Y. et al. Patient-derived organoids predict chemoradiation responses of locally advanced rectal cancer. Cell Stem Cell 26, 17–26.e16 (2020).
de Witte, C. J. et al. Patient-derived ovarian cancer organoids mimic clinical response and exhibit heterogeneous inter- and intrapatient drug responses. Cell Rep. 31, 107762 (2020).
Lorenzo-Martín, L. F. et al. Patient-derived mini-colons enable long-term modeling of tumor–microenvironment complexity. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02301-4 (2024).
Öhlund, D. et al. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214, 579–596 (2017).
Sobrino, A. et al. 3D microtumors in vitro supported by perfused vascular networks. Sci. Rep. 6, 31589 (2016).
Tiriac, H. et al. Organoid profiling identifies common responders to chemotherapy in pancreatic cancer. Cancer Discov. 8, 1112–1129 (2018). This article shows that pharmacotyping using PDTOs correlates with individual patient treatment responses, highlighting the organoid model as a personalized approach to therapy.
Ganesh, K. et al. A rectal cancer organoid platform to study individual responses to chemoradiation. Nat. Med. 25, 1607–1614 (2019).
US National Library of Medicine. clinicaltrials.gov https://clinicaltrials.gov/study/NCT03544047 (2018).
US National Library of Medicine. clinicaltrials.gov https://clinicaltrials.gov/study/NCT04931381 (2022).
US National Library of Medicine. clinicaltrials.gov https://clinicaltrials.gov/study/NCT04859166 (2023).
US National Library of Medicine. clinicaltrials.gov https://clinicaltrials.gov/study/NCT05669586 (2024).
US National Library of Medicine. clinicaltrials.gov https://clinicaltrials.gov/study/NCT03979170 (2024).
Ballet, F. Hepatotoxicity in drug development: detection, significance and solutions. J. Hepatol. 26, 26–36 (1997).
Chen, M. et al. FDA-approved drug labeling for the study of drug-induced liver injury. Drug Discov. Today 16, 697–703 (2011).
Lee, J.-Y. et al. Use of 3D human liver organoids to predict drug-induced phospholipidosis. Int. J. Mol. Sci. 21, 2982 (2020).
Shinozawa, T. et al. High-fidelity drug-induced liver injury screen using human pluripotent stem cell-derived organoids. Gastroenterology 160, 831–846.e810 (2021).
Wu, X., Jiang, D., Yang, Y., Li, S. & Ding, Q. Modeling drug-induced liver injury and screening for anti-hepatofibrotic compounds using human PSC-derived organoids. Cell Regen. 12, 6 (2023).
Wang, Z. et al. Cholangiocyte organoids to study drug-induced injury. Stem Cell Res. Ther. 15, 78 (2024).
Berreur, E. et al. iPSC-hepatocyte organoids as a novel platform to predict AAV gene therapy efficacy. Mol. Ther. Methods Clin. Dev. 33, 101467 (2025).
Hukriede, N. A. et al. Experimental models of acute kidney injury for translational research. Nat. Rev. Nephrol. 18, 277–293 (2022).
Morizane, R. et al. Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat. Biotechnol. 33, 1193–1200 (2015).
Freedman, B. S. et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat. Commun. 6, 8715 (2015).
Czerniecki, S. M. et al. High-throughput screening enhances kidney organoid differentiation from human pluripotent stem cells and enables automated multidimensional phenotyping. Cell Stem Cell 22, 929–940.e924 (2018).
Digby, J. L. M., Vanichapol, T., Przepiorski, A., Davidson, A. J. & Sander, V. Evaluation of cisplatin-induced injury in human kidney organoids. Am. J. Physiol. Ren. Physiol. 318, F971–F978 (2020).
Hale, L. J. et al. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 9, 5167 (2018).
Kumar, S. V. et al. Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells. Development 146, dev172361 (2019).
Lawlor, K. T. et al. Cellular extrusion bioprinting improves kidney organoid reproducibility and conformation. Nat. Mater. 20, 260–271 (2020).
Kroll, K. T. et al. Immune-infiltrated kidney organoid-on-chip model for assessing T cell bispecific antibodies. Proc. Natl Acad. Sci. USA 120, e2305322120 (2023).
Belair, D. G. et al. Human ileal organoid model recapitulates clinical incidence of diarrhea associated with small molecule drugs. Toxicol. In Vitro 68, 104928 (2020).
Rodrigues, D. et al. Unravelling mechanisms of doxorubicin-induced toxicity in 3D human intestinal organoids. Int. J. Mol. Sci. 23, 1286 (2022).
Rodrigues, D. et al. A transcriptomic approach to elucidate the mechanisms of gefitinib-induced toxicity in healthy human intestinal organoids. Int. J. Mol. Sci. 23, 2213 (2022).
Takahashi, Y. et al. Drug cytotoxicity screening using human intestinal organoids propagated with extensive cost-reduction strategies. Sci. Rep. 13, 5407 (2023).
Kourula, S. et al. Intestinal organoids as an in vitro platform to characterize disposition, metabolism, and safety profile of small molecules. Eur. J. Pharm. Sci. 188, 106481 (2023).
Harter, M. F. et al. Analysis of off-tumour toxicities of T-cell-engaging bispecific antibodies via donor-matched intestinal organoids and tumouroids. Nat. Biomed. Eng. 8, 345–360 (2023).
Recaldin, T. et al. Human organoids with an autologous tissue-resident immune compartment. Nature 633, 165–173 (2024).
Cook, D. et al. Lessons learned from the fate of AstraZeneca’s drug pipeline: a five-dimensional framework. Nat. Rev. Drug Discov. 13, 419–431 (2014).
Liu, F., Huang, J. & Liu, Z. Vincristine impairs microtubules and causes neurotoxicity in cerebral organoids. Neuroscience 404, 530–540 (2019).
Huang, J. et al. Tranylcypromine causes neurotoxicity and represses BHC110/LSD1 in human-induced pluripotent stem cell-derived cerebral organoids model. Front. Neurol. 8, 626 (2017).
Scholz, S. et al. Induced pluripotent stem cell-derived brain organoids as potential human model system for chemotherapy induced CNS toxicity. Front. Mol. Biosci. 9, 1006497 (2022).
Destere, A. et al. Drug-induced cardiac toxicity and adverse drug reactions, a narrative review. Therapies 79, 161–172 (2024).
Chen, X. et al. Assessment of doxorubicin toxicity using human cardiac organoids: a novel model for evaluating drug cardiotoxicity. Chem. Biol. Interact. 386, 110777 (2023).
Wu, X., Williams, S., Robidoux, J., Sriramula, S. & Abdel, A. R. Advanced cardiac organoid model for studying doxorubicin-induced cardiotoxicity. Toxicol. Sci. https://doi.org/10.1093/toxsci/kfaf115 (2025).
Richards, D. J. et al. Human cardiac organoids for the modelling of myocardial infarction and drug cardiotoxicity. Nat. Biomed. Eng. 4, 446–462 (2020).
Skardal, A. et al. Drug compound screening in single and integrated multi-organoid body-on-a-chip systems. Biofabrication 12, 025017 (2020).
Mills, R. J. et al. Drug screening in human PSC-cardiac organoids identifies pro-proliferative compounds acting via the mevalonate pathway. Cell Stem Cell 24, 895–907.e896 (2019).
Hoang, P. et al. Engineering spatial-organized cardiac organoids for developmental toxicity testing. Stem Cell Rep. 16, 1228–1244 (2021).
Yin, F. et al. HiPSC-derived multi-organoids-on-chip system for safety assessment of antidepressant drugs. Lab. Chip 21, 571–581 (2021).
Rajan, S. A. P. et al. Probing prodrug metabolism and reciprocal toxicity with an integrated and humanized multi-tissue organ-on-a-chip platform. Acta Biomater. 106, 124–135 (2020).
Homan, K. A. et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262 (2019).
Susa, K. et al. ATP/ADP biosensor organoids for drug nephrotoxicity assessment. Front. Cell Dev. Biol. 11, 1138504 (2023).
Tao, T. et al. Microengineered multi-organoid system from hiPSCs to recapitulate human liver-islet axis in normal and type 2 diabetes. Adv. Sci. 9, e2103495 (2021).
Carvalho, M. R. et al. Gastrointestinal organs and organoids-on-a-chip: advances and translation into the clinics. Biofabrication 15, 042004 (2023).
Mitrofanova, O. et al. Bioengineered human colon organoids with in vivo-like cellular complexity and function. Cell Stem Cell 31, 1175–1186.e1177 (2024).
Altay, G. et al. Self-organized intestinal epithelial monolayers in crypt and villus-like domains show effective barrier function. Sci. Rep. 9, 10140 (2019).
Hinman, S. S., Wang, Y., Kim, R. & Allbritton, N. L. In vitro generation of self-renewing human intestinal epithelia over planar and shaped collagen hydrogels. Nat. Protoc. 16, 352–382 (2020).
Hofer, M., Duque-Correa, M. A. & Lutolf, M. P. Patterned gastrointestinal monolayers with bilateral access as observable models of parasite gut infection. Nat. Biomed. Eng. 9, 1075–1085 (2024).
Nikolaev, M. et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585, 574–578 (2020).
Bardenbacher, M. et al. Permeability analyses and three dimensional imaging of interferon gamma-induced barrier disintegration in intestinal organoids. Stem Cell Res. 35, 101383 (2019).
Leslie, J. L. et al. Persistence and toxin production by clostridium difficile within human intestinal organoids result in disruption of epithelial paracellular barrier function. Infect. Immun. 83, 138–145 (2015).
Watkins, P. B. Drug metabolism by cytochromes P450 in the liver and small bowel. Gastroenterol. Clin. North Am. 21, 511–526 (1992).
Galetin, A. & Houston, J. B. Intestinal and hepatic metabolic activity of five cytochrome P450 enzymes: impact on prediction of first-pass metabolism. J. Pharmacol. Exp. Ther. 318, 1220–1229 (2006).
Baker, T. K. et al. The current status and use of microphysiological systems by the pharmaceutical industry: The International Consortium for innovation and quality microphysiological systems affiliate survey and commentary. Drug Metab. Dispos. 52, 198–209 (2024).
Kratochwil, N. A. et al. Metabolic profiling of human long-term liver models and hepatic clearance predictions from in vitro data using nonlinear mixed-effects modeling. AAPS J. 19, 534–550 (2017).
Myszczyszyn, A. et al. A hollow fiber membrane-based liver organoid-on-a-chip model for examining drug metabolism and transport. Biofabrication 17, 025035 (2025).
Bouwmeester, M. C. et al. Drug metabolism of hepatocyte-like organoids and their applicability in in vitro toxicity testing. Molecules 28, 621 (2023).
Park, E. et al. Development of organoid-based drug metabolism model. Toxicol. Appl. Pharmacol. 385, 114790 (2019).
Lu, W. et al. Crypt organoid culture as an in vitro model in drug metabolism and cytotoxicity studies. Drug. Metab. Dispos. 45, 748–754 (2017).
Zietek, T. et al. Organoids to study intestinal nutrient transport, drug uptake and metabolism – update to the human model and expansion of applications. Front. Bioeng. Biotechnol. 8, 577656 (2020).
Kakni, P., López-Iglesias, C., Truckenmüller, R., Habibović, P. & Giselbrecht, S. PSC-derived intestinal organoids with apical-out orientation as a tool to study nutrient uptake, drug absorption and metabolism. Front. Mol. Biosci. 10, 1102209 (2023).
Musah, S., Bhattacharya, R. & Himmelfarb, J. Kidney disease modeling with organoids and organs-on-chips. Annu. Rev. Biomed. Eng. 26, 383–414 (2024).
Lin, N. Y. C. et al. Renal reabsorption in 3D vascularized proximal tubule models. Proc. Natl Acad. Sci. USA 116, 5399–5404 (2019).
Homan, K. A. et al. Bioprinting of 3D convoluted renal proximal tubules on perfusable chips. Sci. Rep. 6, 34845 (2016).
Bas-Cristóbal Menéndez, A. et al. Creating a kidney organoid-vasculature interaction model using a novel organ-on-chip system. Sci. Rep. 12, 20699 (2022).
Aceves, J. O. et al. 3D proximal tubule-on-chip model derived from kidney organoids with improved drug uptake. Sci. Rep. 12, 14997 (2022).
Schutgens, F. et al. Tubuloids derived from human adult kidney and urine for personalized disease modeling. Nat. Biotechnol. 37, 303–313 (2019).
Tanimizu, N. et al. Generation of functional liver organoids on combining hepatocytes and cholangiocytes with hepatobiliary connections ex vivo. Nat. Commun. 12, 3390 (2021).
Carolina, E. et al. Generation of human iPSC-derived 3D bile duct within liver organoid by incorporating human iPSC-derived blood vessel. Nat. Commun. 15, 7424 (2024).
Vanslambrouck, J. M. et al. Enhanced metanephric specification to functional proximal tubule enables toxicity screening and infectious disease modelling in kidney organoids. Nat. Commun. 13, 5943 (2022).
Shi, M. et al. Human ureteric bud organoids recapitulate branching morphogenesis and differentiate into functional collecting duct cell types. Nat. Biotechnol. 41, 252–261 (2022).
Banan Sadeghian, R. et al. Cells sorted off hiPSC-derived kidney organoids coupled with immortalized cells reliably model the proximal tubule. Commun. Biol. 6, 483 (2023).
Zeng, Z. et al. Generation of patterned kidney organoids that recapitulate the adult kidney collecting duct system from expandable ureteric bud progenitors. Nat. Commun. 12, 3641 (2021).
Ronaldson-Bouchard, K. et al. A multi-organ chip with matured tissue niches linked by vascular flow. Nat. Biomed. Eng. 6, 351–371 (2022).
Skardal, A. et al. Multi-tissue interactions in an integrated three-tissue organ-on-a-chip platform. Sci. Rep. 7, 8837 (2017).
Wimmer, R. A. et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019).
Zhao, Y. et al. Integrating organoids and organ-on-a-chip devices. Clin. Exp. Immunol. 2, 588–608 (2024).
Brandenberg, N. et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat. Biomed. Eng. 4, 863–874 (2020). This representative study showcases an automated organoid platform designed for HTS in drug discovery, incorporating high-content image-based phenotypic analysis.
Wang, Z. et al. Rapid tissue prototyping with micro-organospheres. Stem Cell Rep. 17, 1959–1975 (2022).
Jiang, S. et al. An automated organoid platform with inter-organoid homogeneity and inter-patient heterogeneity. Cell Rep. Med. 1, 100161 (2020).
Hu, Y. et al. Lung cancer organoids analyzed on microwell arrays predict drug responses of patients within a week. Nat. Commun. 12, 2581 (2021).
Joshi, P., Kang, S.-Y., Yu, K.-N., Kothapalli, C. & Lee, M.-Y. High-content imaging of 3D-cultured neural stem cells on a 384-pillar plate for the assessment of cytotoxicity. Toxicol. In Vitro 65, 104765 (2020).
Dong, L. et al. Integrated microfluidic device for drug studies of early C. elegans embryogenesis. Adv. Sci. 5, 1700751 (2018).
Li, L. et al. A high-throughput, open-space and reusable microfluidic chip for combinational drug screening on tumor spheroids. Lab. Chip 21, 3924–3932 (2021).
Bhusal, A. et al. 3D bioprinted hydrogel microfluidic devices for parallel drug screening. ACS Appl. Bio Mater. 5, 4480–4492 (2022).
Haque, M. R. et al. Patient-derived pancreatic cancer-on-a-chip recapitulates the tumor microenvironment. Microsyst. Nanoeng. 8, 36 (2022).
Weng, Y. et al. Self-assembled matrigel-free iPSC-derived liver organoids demonstrate wide-ranging highly differentiated liver functions. Stem Cell 41, 126–139 (2023).
Bircsak, K. M. et al. A 3D microfluidic liver model for high throughput compound toxicity screening in the OrganoPlate®. Toxicology 28, 152667 (2021).
Mori, A. et al. High-throughput bronchus-on-a-chip system for modeling the human bronchus. Sci. Rep. 14, 26248 (2024).
Shi, Q. et al. Co-culture of human primary hepatocytes and nonparenchymal liver cells in the Emulate® liver-chip for the study of drug-induced liver injury. Curr. Protoc. 2, e478 (2022).
Chen, M., Niclis, J. C. & Denham, M. Protocol for generating reproducible miniaturized controlled midbrain organoids. STAR Protoc. 4, 102451 (2023).
Cortés-Llanos, B., Wang, Y., Sims, C. E. & Allbritton, N. L. A technology of a different sort: microraft arrays. Lab. Chip 21, 3204–3218 (2021).
Stern, A. et al. The CellRaft AIR® system: A novel system enabling organoid imaging, identification, and isolation. SLAS Discov. 27, 201–208 (2022).
Seeto, W. J., Tian, Y., Pradhan, S., Minond, D. & Lipke, E. A. Droplet microfluidics-based fabrication of monodisperse poly(ethylene glycol)-fibrinogen breast cancer microspheres for automated drug screening applications. ACS Biomater. Sci. Eng. 8, 3831–3841 (2022).
Beghin, A. et al. Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat. Methods 19, 881–892 (2022).
Park, J.-C. et al. A logical network-based drug-screening platform for Alzheimer’s disease representing pathological features of human brain organoids. Nat. Commun. 12, 280 (2021).
Malakpour-Permlid, A. & Oredsson, S. A novel 3D polycaprolactone high-throughput system for evaluation of toxicity in normoxia and hypoxia. Toxicol. Rep. 8, 627–635 (2021).
Stokar-Regenscheit, N. et al. Complex in vitro model characterization for context of use in toxicologic pathology: use cases by collaborative teams of biologists, bioengineers, and pathologists. Toxicol. Pathol. 52, 123–137 (2024).
Vuille-dit-Bille, E. et al. PEGDA-based HistoBrick for increasing throughput of cryosectioning and immunohistochemistry in organoid and small tissue studies. Sci. Rep. 15, 412 (2025).
Plummer, S. et al. A human iPSC-derived 3D platform using primary brain cancer cells to study drug development and personalized medicine. Sci. Rep. 9, 1407 (2019).
Bell, L. et al. Advanced tissue technologies of blood-brain barrier organoids as high throughput toxicity readouts in drug development. Heliyon 11, e40813 (2025).
Kopec, A. K. et al. Microphysiological systems in early stage drug development: perspectives on current applications and future impact. J. Toxicol. Sci. 46, 99–114 (2021).
Baran, S. W. et al. Perspectives on the evaluation and adoption of complex in vitro models in drug development: Workshop with the FDA and the pharmaceutical industry (IQ MPS Affiliate). Altex 39, 297–314 (2022).
Nelson, C. P. et al. Advancing alternative methods to reduce animal testing. Science 386, 724–726 (2024).
Harrell, A. W. et al. Endeavours made by trade associations, pharmaceutical companies and regulators in the replacement, reduction and refinement of animal experimentation in safety testing of pharmaceuticals. Regul. Toxicol. Pharmacol. 152, 105683 (2024).
Avila, A. M. et al. Gaps and challenges in nonclinical assessments of pharmaceuticals: an FDA/CDER perspective on considerations for development of new approach methodologies. Regul. Toxicol. Pharmacol. 139, 105345 (2023).
US Food and Drug Administration. Implementing alternative methods. fda.gov https://www.fda.gov/science-research/advancing-alternative-methods-fda/implementing-alternative-methods-program (2018).
Fabre, K. et al. Introduction to a manuscript series on the characterization and use of microphysiological systems (MPS) in pharmaceutical safety and ADME applications. Lab. Chip 20, 1049–1057 (2020).
European Commission: Joint Research Centre. Establishing the scientific validity of complex in vitro models—results of a EURL ECVAM survey. op.europa.eu https://doi.org/10.2760/376171 (2021).
Hargrove-Grimes, P., Low, L. A. & Tagle, D. A. Microphysiological systems: what it takes for community adoption. Exp. Biol. Med. 246, 1435–1446 (2021).
OECD. Guidance Document on the Validation and International Acceptance of New or Updated Test Methods for Hazard Assessment. OECD Series on Testing and Assessment, No. 34. https://doi.org/10.1787/e1f1244b-en (OECD Publishing, 2005).
OECD. Guidance Document on Good In Vitro Method Practices (GIVIMP). OECD Series on Testing and Assessment, No. 286. https://doi.org/10.1787/9789264304796-en (OECD Publishing, 2018).
OECD. Guidance Document for Describing Non-Guideline In Vitro Test Methods. OECD Series on Testing and Assessment, No. 211. https://doi.org/10.1787/9789264274730-en (OECD Publishing, 2017).
ICCVAM. Validation, qualification, and regulatory acceptance of new approach methodologies. ntp.niehs.nih.gov https://ntp.niehs.nih.gov/go/ICCVAM-submit (2024).
Krebs, A. et al. Organ-on-chip in development: towards a roadmap for organs-on-chip. Altex 36, 682–699 (2019).
US Food and Drug Administration. Roadmap to reducing animal testing in preclinical safety studies. https://www.fda.gov/media/186092/download (2025).
European Medicines Agency. New approach methodologies EU-IN horizon scanning report. ema.europa.eu https://www.ema.europa.eu/en/documents/report/new-approach-methodologies-eu-horizon-scanning-report_en.pdf (2025).
European Medicines Agency. Regulatory acceptance of new approach methodologies (NAMs) to reduce animal use testing. ema.europa.eu https://www.ema.europa.eu/en/human-regulatory-overview/research-development/ethical-use-animals-medicine-testing/regulatory-acceptance-new-approach-methodologies-nams-reduce-animal-use-testing-related-content-75921 (2025).
Narasimhan, V. et al. Medium-throughput drug screening of patient-derived organoids from colorectal peritoneal metastases to direct personalized therapy. Clin. Cancer Res. 26, 3662–3670 (2020).
Broutier, L. et al. Human primary liver cancer–derived organoid cultures for disease modeling and drug screening. Nat. Med. 23, 1424–1435 (2017).
Driehuis, E. et al. Oral mucosal organoids as a potential platform for personalized cancer therapy. Cancer Discov. 9, 852–871 (2019).
Derouet, M. F. et al. Towards personalized induction therapy for esophageal adenocarcinoma: organoids derived from endoscopic biopsy recapitulate the pre-treatment tumor. Sci. Rep. 10, 14514 (2020).
Li, X. et al. Organoid cultures recapitulate esophageal adenocarcinoma heterogeneity providing a model for clonality studies and precision therapeutics. Nat. Commun. 9, 2983 (2018).
Li, Z. et al. Patient-derived renal cell carcinoma organoids for personalized cancer therapy. Clin. Transl. Med. 12, e970 (2022).
Loong, H. H. F. et al. Patient-derived tumor organoid predicts drugs response in glioblastoma: A step forward in personalized cancer therapy? J. Clin. Neurosci. 78, 400–402 (2020).
Velazquez, J. J. et al. Gene regulatory network analysis and engineering directs development and vascularization of multilineage human liver organoids. Cell Syst. 12, 41–55.e11 (2021).
Byeon, J. H., Jung, D. J., Han, H.-J., Son, W.-C. & Jeong, G. S. Fast formation and maturation enhancement of human liver organoids using a liver-organoid-on-a-chip. Front. Cell Dev. Biol. 12, 1452485 (2024).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Noel, G. et al. A primary human macrophage-enteroid co-culture model to investigate mucosal gut physiology and host-pathogen interactions. Sci. Rep. 7, 45270 (2017).
Sebrell, T. A. et al. A novel gastric spheroid co-culture model reveals chemokine-dependent recruitment of human dendritic cells to the gastric epithelium. Cell. Mol. Gastroenterol. Hepatol. 8, 157–171.e153 (2019).
de Hoyos-Vega, J. M. et al. Modeling gut neuro-epithelial connections in a novel microfluidic device. Microsyst. Nanoeng. 9, 144 (2023).
Eicher, A. K. et al. Functional human gastrointestinal organoids can be engineered from three primary germ layers derived separately from pluripotent stem cells. Cell Stem Cell 29, 36–51.e36 (2022).
Wijnakker, J. J. A. P. M. et al. Integrin-activating Yersinia protein Invasin sustains long-term expansion of primary epithelial cells as 2D organoid sheets. Proc. Natl Acad. Sci. USA 122, e2420595121 (2025).
Herpers, B. et al. Functional patient-derived organoid screenings identify MCLA-158 as a therapeutic EGFR × LGR5 bispecific antibody with efficacy in epithelial tumors. Nat. Cancer 3, 418–436 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06496178 (2025).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06525220 (2025).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT03526835 (2025).
Acknowledgements
The authors thank S. Fowler and A. Schneider from Roche Pharma Research and Early Development (pRED) for their thorough review and insightful suggestions. This work was supported by Oncode Accelerator, a Dutch National Growth Fund project under the grant number NGFOP2201; the Netherlands Organ-on-Chip Initiative, an NWO Gravitation project (grant number 024.003.001) funded by the Ministry of Education, Culture and Science of the government of the Netherlands; and the Oncode Institute (partly financed by the Dutch Cancer Society).
Author information
Authors and Affiliations
Contributions
D.W., R.V., and N.S.-R. researched data for article. All authors provided substantial contribution to discussion of content, wrote and reviewed and edited the article before submission.
Corresponding author
Ethics declarations
Competing interests
H.C. was the head of Pharma Research and Early Development (pRED) at Roche, Basel, and holds several patents related to organoid technology. His full disclosure can be found at: www.uu.nl/staff/JCClevers/Additional functions. R.V. and N.S.-R. are employees and stockholders of F. Hoffmann-La Roche Ldt.
Peer review
Peer review information
Nature Reviews Drug Discovery thanks Davide Gianni and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, D., Villenave, R., Stokar-Regenscheit, N. et al. Human organoids as 3D in vitro platforms for drug discovery: opportunities and challenges. Nat Rev Drug Discov (2025). https://doi.org/10.1038/s41573-025-01317-y
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41573-025-01317-y