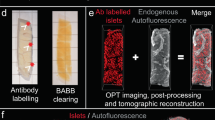
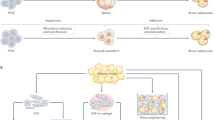
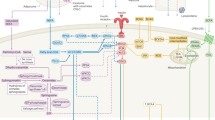
Abstract
Our understanding of diabetes mellitus has benefited from a combination of clinical investigations and work in model organisms and cell lines. Organoid models for a wide range of tissues are emerging as an additional tool enabling the study of diabetes mellitus. The applications for organoid models include studying human pancreatic cell development, pancreatic physiology, the response of target organs to pancreatic hormones and how glucose toxicity can affect tissues such as the blood vessels, retina, kidney and nerves. Organoids can be derived from human tissue cells or pluripotent stem cells and enable the production of human cell assemblies mimicking human organs. Many organ mimics relevant to diabetes mellitus are already available, but only a few relevant studies have been performed. We discuss the models that have been developed for the pancreas, liver, kidney, nerves and vasculature, how they complement other models, and their limitations. In addition, as diabetes mellitus is a multi-organ disease, we highlight how a merger between the organoid and bioengineering fields will provide integrative models.
Key points
-
Pancreas and islet organoids enable the study of monogenic forms of diabetes mellitus, notably neonatal diabetes mellitus.
-
For type 1 diabetes mellitus, platforms mixing patient-relevant islets and immune cells are expanding.
-
Organoid models are emerging that combine cell types derived from multiple organs relevant to diabetes mellitus.
-
A merger between bioengineering techniques and organoid technology is improving functional and screening assays.
-
For assays of organ interactions, organ-on-a-chip platforms are moving from cell lines to organoids.
-
While organoid models for diabetes mellitus complications are being added to the toolbox, the long periods over which complications develop mean that studying these complications in vitro will be challenging.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Nauck, M. A., Wefers, J. & Meier, J. J. Treatment of type 2 diabetes: challenges, hopes, and anticipated successes. Lancet Diabetes Endocrinol. 9, 525–544 (2021).
Huch, M., Knoblich, J. A., Lutolf, M. P. & Martinez-Arias, A. The hope and the hype of organoid research. Development 144, 938–941 (2017). This comprehensive review on organoids delves into the history and philosophy of the organoid field.
Marsee, A. et al. Building consensus on definition and nomenclature of hepatic, pancreatic, and biliary organoids. Cell Stem Cell 28, 816–832 (2021).
Tsakmaki, A., Fonseca Pedro, P. & Bewick, G. A. Diabetes through a 3D lens: organoid models. Diabetologia 63, 1093–1102 (2020).
Hopcroft, D. W., Mason, D. R. & Scott, R. S. Insulin secretion from perifused rat pancreatic pseudoislets. Vitr. Cell. Dev. Biol. 21, 421–427 (1985).
Halban, P. A., Powers, S. L., George, K. L. & Bonner-Weir, S. Spontaneous reassociation of dispersed adult rat pancreatic islet cells into aggregates with three-dimensional architecture typical of native islets. Diabetes 36, 783–790 (1987).
Hilderink, J. et al. Controlled aggregation of primary human pancreatic islet cells leads to glucose-responsive pseudoislets comparable to native islets. J. Cell. Mol. Med. 19, 1836–1846 (2015).
Velazco-Cruz, L., Goedegebuure, M. M. & Millman, J. R. Advances toward engineering functionally mature human pluripotent stem cell-derived β cells. Front. Bioeng. Biotechnol. 8, 786 (2020).
Brusko, T. M., Russ, H. A. & Stabler, C. L. Strategies for durable β cell replacement in type 1 diabetes. Science 373, 516–522 (2021).
Frum, T. & Spence, J. R. hPSC-derived organoids: models of human development and disease. J. Mol. Med. 99, 463–473 (2021).
Beltrand, J. et al. Neonatal diabetes mellitus. Front. Pediatr. 8, 602 (2020).
O’Hara, S. E., Gembus, K. M. & Nicholas, L. M. Understanding the long-lasting effects of fetal nutrient restriction versus exposure to an obesogenic diet on islet-cell mass and function. Metabolites 11, 514 (2021).
Larsen, H. L. & Grapin-Botton, A. The molecular and morphogenetic basis of pancreas organogenesis. Semin. Cell Dev. Biol. 66, 51–68 (2017).
Petersen, M. B. K., Gonçalves, C. A. C., Kim, Y. H. & Grapin-Botton, A. Recapitulating and deciphering human pancreas development from human pluripotent stem cells in a dish. Curr. Top. Dev. Biol. 129, 143–190 (2018).
Sugiyama, T. et al. Reconstituting pancreas development from purified progenitor cells reveals genes essential for islet differentiation. Proc. Natl Acad. Sci. USA 110, 12691–12696 (2013).
Greggio, C. et al. Artificial three-dimensional niches deconstruct pancreas development in vitro. Development 140, 4452–4462 (2013). This article reports on the first pancreas organoid models, which remain the most complex organoid system in terms of architecture and cell diversity.
Bonfanti, P. et al. Ex vivo expansion and differentiation of human and mouse fetal pancreatic progenitors are modulated by epidermal growth factor. Stem Cell Dev. 24, 1766–1778 (2015).
Bakhti, M. et al. Establishment of a high-resolution 3D modeling system for studying pancreatic epithelial cell biology in vitro. Mol. Metab. 30, 16–29 (2019).
Scavuzzo, M. A., Yang, D. & Borowiak, M. Organotypic pancreatoids with native mesenchyme develop insulin producing endocrine cells. Sci. Rep. 7, 10810 (2017).
Gonçalves, C. A. et al. A 3D system to model human pancreas development and its reference single-cell transcriptome atlas identify signaling pathways required for progenitor expansion. Nat. Commun. 12, 3144 (2021).
Loomans, C. J. M. et al. Expansion of adult human pancreatic tissue yields organoids harboring progenitor cells with endocrine differentiation potential. Stem Cell Rep. 10, 712–724 (2018).
Dahl-Jensen, S. B. et al. Deconstructing the principles of ductal network formation in the pancreas. PLoS Biol. 16, e2002842 (2018).
Scavuzzo, M. A., Teaw, J., Yang, D. & Borowiak, M. Generation of scaffold-free, three-dimensional insulin expressing pancreatoids from mouse pancreatic progenitors in vitro. J. Vis. Exp. 2018, e57599 (2018).
Nair, G. G., Tzanakakis, E. S. & Hebrok, M. Emerging routes to the generation of functional β-cells for diabetes mellitus cell therapy. Nat. Rev. Endocrinol. 16, 506–518 (2020).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133 (2014).
Nostro, M. C. et al. Stage-specific signaling through TGFβ family members and WNT regulates patterning and pancreatic specification of human pluripotent stem cells. Development 138, 861–871 (2011).
Pagliuca, F. W. et al. Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439 (2014).
Russ, H. A. et al. Controlled induction of human pancreatic progenitors produces functional beta‐like cells in vitro. EMBO J. 34, 1759–1772 (2015).
Nair, G. G. et al. Recapitulating endocrine cell clustering in culture promotes maturation of human stem-cell-derived β cells. Nat. Cell Biol. 21, 263–274 (2019). The innovation of this article consisted in sorting endocrine cells produced from PSCs to form islets of enhanced functionality compared with islets that contain undifferentiated progenitors.
Velazco-Cruz, L. et al. Acquisition of dynamic function in human stem cell-derived β cells. Stem Cell Rep. 12, 351–365 (2019).
Toyoda, T. et al. Cell aggregation optimizes the differentiation of human ESCs and iPSCs into pancreatic bud-like progenitor cells. Stem Cell Res. 14, 185–197 (2015).
Jiang, J. et al. Generation of insulin‐producing islet‐like clusters from human embryonic. Stem Cell. Stem Cells 25, 1940–1953 (2007).
Zhu, S. et al. Human pancreatic beta-like cells converted from fibroblasts. Nat. Commun. 7, 10080 (2016).
Balboa, D. et al. Functional, metabolic and transcriptional maturation of human pancreatic islets derived from stem cells. Nat. Biotechnol. 40, 1042–1055 (2022). The current highest quality assessment standards for benchmarking in vitro-produced islets.
Hohwieler, M. et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 66, 473–486 (2017).
Wesolowska-Andersen, A. et al. Analysis of differentiation protocols defines a common pancreatic progenitor molecular signature and guides refinement of endocrine differentiation. Stem Cell Rep. 14, 138–153 (2020).
Wang, X. & Ye, K. Three-dimensional differentiation of embryonic stem cells into islet-like insulin-producing clusters. Tissue Eng. Part A 15, 1941–1952 (2009).
Saito, H., Takeuchi, M., Chida, K. & Miyajima, A. Generation of glucose-responsive functional islets with a three-dimensional structure from mouse fetal pancreatic cells and iPS cells in vitro. PLoS ONE 6, e28209 (2011).
Braverman-Gross, C. & Benvenisty, N. Modeling maturity onset diabetes of the young in pluripotent stem cells: challenges and achievements. Front. Endocrinol. 12, 20 (2021).
Riddle, M. C. et al. Monogenic diabetes: from genetic insights to population-based precision in care. Reflections from a diabetes care editors’ expert forum. Diabetes Care 43, 3117–3128 (2020).
Balboa, D., Iworima, D. G. & Kieffer, T. J. Human pluripotent stem cells to model islet defects in diabetes. Front. Endocrinol. 12, 149 (2021).
Abdelalim, E. M. Modeling different types of diabetes using human pluripotent stem cells. Cell. Mol. Life Sci. 78, 2459–2483 (2021).
Ma, S. et al. β Cell replacement after gene editing of a neonatal diabetes-causing mutation at the insulin locus. Stem Cell Rep. 11, 1407–1415 (2018).
Balboa, D. et al. Insulin mutations impair beta-cell development in a patient-derived iPSC model of neonatal diabetes. Elife 7, e38519 (2018).
Panova, A. V. et al. Aberrant splicing of INS impairs beta-cell differentiation and proliferation by ER stress in the isogenic iPSC model of neonatal diabetes. Int. J. Mol. Sci. 23, 8824 (2022).
De Franco, E. et al. YIPF5 mutations cause neonatal diabetes and microcephaly through endoplasmic reticulum stress. J. Clin. Invest. 130, 6338–6353 (2020).
Saarimäki-Vire, J. et al. An activating STAT3 mutation causes neonatal diabetes through premature induction of pancreatic differentiation. Cell Rep. 19, 281–294 (2017).
Zhou, Z. et al. A missense KCNQ1 mutation impairs insulin secretion in neonatal diabetes. bioRxiv https://doi.org/10.1101/2021.08.24.457485v1 (2021).
Wang, X. et al. Point mutations in the PDX1 transactivation domain impair human β-cell development and function. Mol. Metab. 24, 80–97 (2019).
Zhu, Z. et al. Genome editing of lineage determinants in human pluripotent stem cells reveals mechanisms of pancreatic development and diabetes. Cell Stem Cell 18, 755–768 (2016).
Zhang, X. et al. A comprehensive structure-function study of neurogenin3 disease-causing alleles during human pancreas and intestinal organoid development. Dev. Cell 50, 367–380.e7 (2019).
Philippi, A. et al. Mutations and variants of ONECUT1 in diabetes. Nat. Med. 27, 1928–1940 (2021).
Teo, A. K. K. et al. Early developmental perturbations in a human stem cell model of MODY5/HNF1B pancreatic hypoplasia. Stem Cell Rep. 6, 357–367 (2016).
Vethe, H. et al. Probing the missing mature β-cell proteomic landscape in differentiating patient iPSC-derived cells. Sci. Rep. 7, 4780 (2017).
Braverman-Gross, C. et al. Derivation and molecular characterization of pancreatic differentiated MODY1-iPSCs. Stem Cell Res. 31, 16–26 (2018).
Yabe, S. G. et al. Expression of mutant mRNA and protein in pancreatic cells derived from MODY3- iPS cells. PLoS ONE 14, e0217110 (2019).
Cardenas-Diaz, F. L. et al. Modeling monogenic diabetes using human ESCs reveals developmental and metabolic deficiencies caused by mutations in HNF1A. Cell Stem Cell 25, 273–289.e5 (2019).
González, B. J. et al. Reduced calcium levels and accumulation of abnormal insulin granules in stem cell models of HNF1A deficiency. Commun. Biol. 5, 779 (2022).
Cujba, A.-M. et al. An HNF1α truncation associated with maturity-onset diabetes of the young impairs pancreatic progenitor differentiation by antagonizing HNF1β function. Cell Rep. 38, 110425 (2022).
DeForest, N. & Majithia, A. R. Genetics of type 2 diabetes: implications from large-scale studies. Curr. Diab. Rep. 22, 227–235 (2022).
Veres, A. et al. Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373 (2019).
Augsornworawat, P., Maxwell, K. G., Velazco-Cruz, L. & Millman, J. R. Single-cell transcriptome profiling reveals β cell maturation in stem cell-derived islets after transplantation. Cell Rep. 32, 108067 (2020).
Davis, J. C. et al. Glucose response by stem cell-derived β cells in vitro is inhibited by a bottleneck in glycolysis. Cell Rep. 31, 107623 (2020).
Basford, C. L. et al. The functional and molecular characterisation of human embryonic stem cell-derived insulin-positive cells compared with adult pancreatic beta cells. Diabetologia 55, 358–371 (2012).
Alvarez-Dominguez, J. R. & Melton, D. A. Cell maturation: hallmarks, triggers, and manipulation. Cell 185, 235–249 (2022).
Helman, A. et al. A nutrient-sensing transition at birth triggers glucose-responsive insulin secretion. Cell Metab. 31, 1004–1016.e5 (2020).
Wang, D. et al. Long-term expansion of pancreatic islet organoids from resident Procr+ progenitors. Cell 180, 1198–1211.e19 (2020). This article suggests that there are progenitor populations in islets that can seed organoids.
Domínguez-Bendala, J., Qadir, M. M. F. & Pastori, R. L. Pancreatic progenitors: there and back again. Trends Endocrinol. Metab. 30, 4–11 (2019).
Huch, M. et al. Unlimited in vitro expansion of adult bi-potent pancreas progenitors through the Lgr5/R-spondin axis. EMBO J. 32, 2708–2721 (2013). This article reports on the first adult pancreatic duct-derived spheres and their potential to produce endocrine cells.
Rezanejad, H. et al. Heterogeneity of SOX9 and HNF1β in pancreatic ducts is dynamic. Stem Cell Rep. 10, 725–738 (2018).
Beşikcioğlu, H. E. et al. Organoids: questions and answers. STAR Protocols https://star-protocols.cell.com/categories/organoids/qa# (2022).
Montesano, R., Mouron, P., Amherdt, M. & Orci, L. Collagen matrix promotes reorganization of pancreatic endocrine cell monolayers into islet-like organoids. J. Cell Biol. 97, 935–939 (1983).
Oberg-Welsh, C. Long-term culture in matrigel enhances the insulin secretion of fetal porcine islet-like cell clusters in vitro. Pancreas 22, 157–163 (2001).
Wang, W., Jin, S. & Ye, K. Development of islet organoids from H9 human embryonic stem cells in biomimetic 3D scaffolds. Stem Cell Dev. 26, 394–404 (2017).
Kozlowski, M. T., Crook, C. J. & Ku, H. T. Towards organoid culture without Matrigel. Commun. Biol. 4, 1387 (2021).
Candiello, J. et al. 3D heterogeneous islet organoid generation from human embryonic stem cells using a novel engineered hydrogel platform. Biomaterials 177, 27–39 (2018).
Jiang, K. et al. 3-D physiomimetic extracellular matrix hydrogels provide a supportive microenvironment for rodent and human islet culture. Biomaterials 198, 37–48 (2019).
Mohammed, J. S., Wang, Y., Harvat, T. A., Oberholzer, J. & Eddington, D. T. Microfluidic device for multimodal characterization of pancreatic islets. Lab Chip 9, 97–106 (2009). One of the first pancreas-on-a-chip systems to assess human islet functionality after isolation.
Abadpour, S. et al. Pancreas-on-a-chip technology for transplantation applications. Curr. Diab. Rep. 20, 72 (2020).
Glieberman, A. L. et al. Synchronized stimulation and continuous insulin sensing in a microfluidic human islet on a chip designed for scalable manufacturing. Lab Chip 19, 2993–3010 (2019). This islet-on-a-chip system includes in situ automated glucose stimulation and measurement inside the chip.
Lee, S. H. et al. Microphysiological analysis platform of pancreatic islet β-cell spheroids. Adv. Healthc. Mater. 7, 1701111 (2018).
Jun, Y. et al. In vivo-mimicking microfluidic perfusion culture of pancreatic islet spheroids. Sci. Adv. 5, eaax4520 (2019).
Essaouiba, A. et al. Microwell-based pancreas-on-chip model enhances genes expression and functionality of rat islets of Langerhans. Mol. Cell. Endocrinol. 514, 110892 (2020).
Tao, T. et al. Engineering human islet organoids from iPSCs using an organ-on-chip platform. Lab Chip 19, 948–958 (2019).
Liu, H. et al. A droplet microfluidic system to fabricate hybrid capsules enabling stem cell organoid engineering. Adv. Sci. 7, 1903739 (2020).
Zbinden, A. et al. Non-invasive marker-independent high content analysis of a microphysiological human pancreas-on-a-chip model. Matrix Biol. 85–86, 205–220 (2020).
Patel, S. N. et al. Organoid microphysiological system preserves pancreatic islet function within 3D matrix. Sci. Adv. 7, eaba5515 (2021).
Walker, J. T. et al. Integrated human pseudoislet system and microfluidic platform demonstrate differences in GPCR signaling in islet cells. JCI Insight 5, e137017 (2020).
Kumar, S. A., Delgado, M., Mendez, V. E. & Joddar, B. Applications of stem cells and bioprinting for potential treatment of diabetes. World J. Stem Cell 11, 13–32 (2019).
Amin, J. et al. A simple, reliable method for high-throughput screening for diabetes drugs using 3D β-cell spheroids. J. Pharmacol. Toxicol. Methods 82, 83–89 (2016).
Yang, K. et al. A 3D culture platform enables development of zinc-binding prodrugs for targeted proliferation of β cells. Sci. Adv. 6, 3207–3225 (2020).
Penko, D. et al. Incorporation of endothelial progenitor cells into mosaic pseudoislets. Islets 3, 73–79 (2011).
Spelios, M. G., Afinowicz, L. A., Tipon, R. C. & Akirav, E. M. Human EndoC-βH1 β-cells form pseudoislets with improved glucose sensitivity and enhanced GLP-1 signaling in the presence of islet-derived endothelial cells. Am. J. Physiol. Metab. 314, E512–E521 (2018).
Urbanczyk, M., Zbinden, A., Layland, S. L., Duffy, G. & Schenke-Layland, K. Controlled heterotypic pseudo-islet assembly of human β-cells and human umbilical vein endothelial cells using magnetic levitation. Tissue Eng. Part. A 26, 387–399 (2020).
Augsornworawat, P., Velazco-Cruz, L., Song, J. & Millman, J. R. A hydrogel platform for in vitro three dimensional assembly of human stem cell-derived islet cells and endothelial cells. Acta Biomater. 97, 272–280 (2019).
Seo, H., Son, J. & Park, J.-K. Controlled 3D co-culture of beta cells and endothelial cells in a micropatterned collagen sheet for reproducible construction of an improved pancreatic pseudo-tissue. Apl. Bioeng. 4, 046103 (2020).
Nalbach, L. et al. Improvement of islet transplantation by the fusion of islet cells with functional blood vessels. EMBO Mol. Med. 13, e12616 (2021).
Aghazadeh, Y. et al. Microvessels support engraftment and functionality of human islets and hESC-derived pancreatic progenitors in diabetes models. Cell Stem Cell 28, 1936–1949.e8 (2021).
Rambøl, M. H., Han, E. & Niklason, L. E. Microvessel network formation and interactions with pancreatic islets in three-dimensional chip cultures. Tissue Eng. Part. A 26, 556–568 (2020).
Palikuqi, B. et al. Adaptable haemodynamic endothelial cells for organogenesis and tumorigenesis. Nature 585, 426–432 (2020).
Soltanian, A. et al. Generation of functional human pancreatic organoids by transplants of embryonic stem cell derivatives in a 3D‐printed tissue trapper. J. Cell. Physiol. 234, 9564–9576 (2019).
Takebe, T. et al. Vascularized and complex organ buds from diverse tissues via mesenchymal cell-driven condensation. Cell Stem Cell 16, 556–565 (2015). This article sets up a method to make organoids by mixing epithelial, mesenchymal and vascular cells.
Takahashi, Y., Sekine, K., Kin, T., Takebe, T. & Taniguchi, H. Self-condensation culture enables vascularization of tissue fragments for efficient therapeutic transplantation. Cell Rep. 23, 1620–1629 (2018).
Takahashi, Y., Takebe, T. & Taniguchi, H. Methods for generating vascularized islet‐like organoids via self‐condensation. Curr. Protoc. Stem Cell Biol. 45, e49 (2018).
Jiao, A. et al. Simulated cholinergic reinnervation of β (INS-1) cells: antidiabetic utility of heterotypic pseudoislets containing β cell and cholinergic cell. Int. J. Endocrinol. https://doi.org/10.1155/2018/1505307 (2018).
Besikcioglu, H. E. et al. Innervated mouse pancreas organoids as an ex vivo model to study pancreatic neuropathy in pancreatic cancer. STAR Protoc. 2, 100935 (2021).
Chiou, J. et al. Interpreting type 1 diabetes risk with genetics and single-cell epigenomics. Nature 594, 398–402 (2021).
Kahraman, S. et al. Abnormal exocrine–endocrine cell cross-talk promotes β-cell dysfunction and loss in MODY8. Nat. Metab. 4, 76–89 (2022).
Overton, D. L. & Mastracci, T. L. Exocrine-endocrine crosstalk: the influence of pancreatic cellular communications on organ growth, function and disease. Front. Endocrinol. 13, 1010 (2022).
Huang, L. et al. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell 28, 1090–1104.e6 (2021).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371 (2015).
Khosravi-Maharlooei, M. et al. Modeling human T1D-associated autoimmune processes. Mol. Metab. 56, 101417 (2021).
Skyler, J. S. Hope vs hype: where are we in type 1 diabetes? Diabetologia 61, 509–516 (2018).
Concannon, P., Rich, S. S. & Nepom, G. T. Genetics of type 1A diabetes. N. Engl. J. Med. 360, 1646–1654 (2009).
Joshi, K. et al. Modeling type 1 diabetes using pluripotent stem cell technology. Front. Endocrinol. 12, 260 (2021).
Maehr, R. et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl Acad. Sci. USA 106, 15768–15773 (2009).
Thatava, T. et al. Intrapatient variations in type 1 diabetes-specific iPS cell differentiation into insulin-producing cells. Mol. Ther. 21, 228–239 (2013).
Millman, J. R. et al. Generation of stem cell-derived β-cells from patients with type 1 diabetes. Nat. Commun. 7, 11463 (2016).
Hosokawa, Y. et al. Insulin-producing cells derived from ‘induced pluripotent stem cells’ of patients with fulminant type 1 diabetes: vulnerability to cytokine insults and increased expression of apoptosis-related genes. J. Diabetes Investig. 9, 481–493 (2018).
Leite, N. C. et al. Modeling type 1 diabetes in vitro using human pluripotent stem cells. Cell Rep. 32, 107894 (2020).
Culina, S. et al. Islet-reactive CD8+ T cell frequencies in the pancreas, but not in blood, distinguish type 1 diabetic patients from healthy donors. Sci. Immunol. 3, 4013 (2018).
Armitage, L. H. et al. Use of induced pluripotent stem cells to build isogenic systems and investigate type 1 diabetes. Front. Endocrinol. 12, 1376 (2021).
Joshi, K. et al. Induced pluripotent stem cell macrophages present antigen to proinsulin-specific T cell receptors from donor-matched islet-infiltrating T cells in type 1 diabetes. Diabetologia 62, 2245–2251 (2019).
Amin, S. et al. Discovery of a drug candidate for GLIS3-associated diabetes. Nat. Commun. 9, 2681 (2018).
Prior, N., Inacio, P. & Huch, M. Liver organoids: from basic research to therapeutic applications. Gut 68, 2228–2237 (2019).
Thompson, W. L. & Takebe, T. Human liver model systems in a dish. Dev. Growth Differ. 63, 47–58 (2021).
Camp, J. G. et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 546, 533–538 (2017).
Ardalani, H. et al. 3-D culture and endothelial cells improve maturity of human pluripotent stem cell-derived hepatocytes. Acta Biomater. 95, 371–381 (2019).
Zabulica, M. et al. Guide to the assessment of mature liver gene expression in stem cell-derived hepatocytes. Stem Cell Dev. 28, 907–919 (2019).
Thompson, W. L. & Takebe, T. Generation of multi-cellular human liver organoids from pluripotent stem cells. in. Methods Cell Biol. 159, 47–68 (2020).
Takebe, T. et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484 (2013).
Prior, N. et al. Lgr5+ stem/progenitor cells reside at the apex of a heterogeneous embryonic hepatoblast pool. Development 146, dev174557 (2019).
Huch, M. et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 160, 299–312 (2015).
Peng, W. C. et al. Inflammatory cytokine TNFα promotes the long-term expansion of primary hepatocytes in 3D culture. Cell 175, 1607–1619.e15 (2018). While liver ductal organoids had been produced before, this article together with Hu et al. (2018) reported the first hepatocyte organoid systems.
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606.e19 (2018).
Kruitwagen, H. S. et al. Long-term adult feline liver organoid cultures for disease modeling of hepatic steatosis. Stem Cell Rep. 8, 822–830 (2017).
Ouchi, R. et al. Modeling steatohepatitis in humans with pluripotent stem cell-derived organoids. Cell Metab. 30, 374–384.e6 (2019).
Fair, K. L., Colquhoun, J. & Hannan, N. R. F. Intestinal organoids for modelling intestinal development and disease. Philos. Trans. R. Soc. B Biol. Sci. 373, 20170217 (2018).
Sato, T. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). This article can be considered one of the founding papers of the organoid field, reporting on the in vitro production of 3D organoids from intestinal stem cells.
Spence, J. R. et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109 (2011).
Zietek, T., Rath, E., Haller, D. & Daniel, H. Intestinal organoids for assessing nutrient transport, sensing and incretin secretion. Sci. Rep. 5, 16831 (2015).
Nguyen, D. T. T., van Noort, D., Jeong, I.-K. & Park, S. Endocrine system on chip for a diabetes treatment model. Biofabrication 9, 015021 (2017).
Goldspink, D. A. et al. Labeling and characterization of human GLP-1-secreting L-cells in primary ileal organoid culture. Cell Rep. 31, 107833 (2020).
Vandenburgh, H. et al. Tissue-engineered skeletal muscle organoids for reversible gene therapy. Hum. Gene Ther. 7, 2195–2200 (1996).
Jalal, S., Dastidar, S. & Tedesco, F. S. Advanced models of human skeletal muscle differentiation, development and disease: three-dimensional cultures, organoids and beyond. Curr. Opin. Cell Biol. 73, 92–104 (2021).
Maffioletti, S. M. et al. Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep. 23, 899–908 (2018).
Rothman, D. L. et al. Decreased muscle glucose transport/phosphorylation is an early defect in the pathogenesis of non-insulin-dependent diabetes mellitus. Proc. Natl Acad. Sci. USA 92, 983–987 (1995).
Iovino, S., Burkart, A. M., Warren, L., Patti, M. E. & Kahn, C. R. Myotubes derived from human-induced pluripotent stem cells mirror in vivo insulin resistance. Proc. Natl Acad. Sci. USA 113, 1889–1894 (2016).
Batista, T. M. et al. A cell-autonomous signature of dysregulated protein phosphorylation underlies muscle insulin resistance in type 2 diabetes. Cell Metab. 32, 844–859.e5 (2020).
Ghaben, A. L. & Scherer, P. E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell Biol. 20, 242–258 (2019).
Klingelhutz, A. J. et al. Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery. Sci. Rep. 8, 523 (2018).
Taylor, J. et al. Generation of immune cell containing adipose organoids for in vitro analysis of immune metabolism. Sci. Rep. 10, 21104 (2020).
Muller, S. et al. Human adipose stromal-vascular fraction self-organizes to form vascularized adipose tissue in 3D cultures. Sci. Rep. 9, 7250 (2019).
Liu, Y. et al. Adipose-on-a-chip: a dynamic microphysiological in vitro model of the human adipose for immune-metabolic analysis in type II diabetes. Lab Chip 19, 241–253 (2019).
McCarthy, M. et al. Fat-on-a-chip models for research and discovery in obesity and its metabolic comorbidities. Tissue Eng. Part. B Rev. 26, 586–595 (2020).
Bauer, S. et al. Functional coupling of human pancreatic islets and liver spheroids on-a-chip: towards a novel human ex vivo type 2 diabetes model. Sci. Rep. 7, 14620 (2017).
Miller, P. G. & Shuler, M. L. Design and demonstration of a pumpless 14 compartment microphysiological system. Biotechnol. Bioeng. 113, 2213–2227 (2016).
Edington, C. D. et al. Interconnected microphysiological systems for quantitative biology and pharmacology studies. Sci. Rep. 8, 4530 (2018).
Rogal, J., Zbinden, A., Schenke-Layland, K. & Loskill, P. Stem-cell based organ-on-a-chip models for diabetes research. Adv. Drug Deliv. Rev. 140, 101–128 (2019).
Tao, T. et al. Microengineered multi‐organoid system from hiPSCs to recapitulate human liver‐islet axis in normal and type 2 diabetes. Adv. Sci. 9, 2103495 (2022).
Wimmer, R. A. et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 565, 505–510 (2019).
Nguyen, J., Lin, Y.-Y. & Gerecht, S. The next generation of endothelial differentiation: tissue-specific ECs. Cell Stem Cell 28, 1188–1204 (2021).
Gao, G. et al. Construction of a novel in vitro atherosclerotic model from geometry‐tunable artery equivalents engineered via in‐bath coaxial cell printing. Adv. Funct. Mater. 31, 2008878 (2021).
Zhang, S., Wan, Z. & Kamm, R. D. Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature. Lab Chip 21, 473–488 (2021).
Drawnel, F. M. et al. Disease modeling and phenotypic drug screening for diabetic cardiomyopathy using human induced pluripotent stem cells. Cell Rep. 9, 810–820 (2014).
Azushima, K., Gurley, S. B. & Coffman, T. M. Modelling diabetic nephropathy in mice. Nat. Rev. Nephrol. 14, 48–56 (2018).
Garreta, E. et al. A diabetic milieu increases ACE2 expression and cellular susceptibility to SARS-CoV-2 infections in human kidney organoids and patient cells. Cell Metab. 34, 857–873.e9 (2022).
Nishinakamura, R. Human kidney organoids: progress and remaining challenges. Nat. Rev. Nephrol. 15, 613–624 (2019).
Buzhor, E. et al. Kidney spheroids recapitulate tubular organoids leading to enhanced tubulogenic potency of human kidney-derived cells. Tissue Eng. Part A 17, 2305–2319 (2011).
Garreta, E. et al. Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat. Mater. 18, 397–405 (2019).
Kaisto, S. et al. Optimization of renal organoid and organotypic culture for vascularization, extended development, and improved microscopy imaging. J. Vis. Exp. 2020, e60995 (2020).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Ryan, A. R. et al. Vascular deficiencies in renal organoids and ex vivo kidney organogenesis. Dev. Biol. 477, 98–116 (2021).
Homan, K. A. et al. Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat. Methods 16, 255–262 (2019).
Lin, N. Y. C. et al. Renal reabsorption in 3D vascularized proximal tubule models. Proc. Natl Acad. Sci. USA 116, 5399–5404 (2019).
Sloan, G., Selvarajah, D. & Tesfaye, S. Pathogenesis, diagnosis and clinical management of diabetic sensorimotor peripheral neuropathy. Nat. Rev. Endocrinol. 17, 400–420 (2021).
Pereira, J. D. et al. Human sensorimotor organoids derived from healthy and amyotrophic lateral sclerosis stem cells form neuromuscular junctions. Nat. Commun. 12, 4744 (2021).
Lee, J. et al. Hair-bearing human skin generated entirely from pluripotent stem cells. Nature 582, 399–404 (2020).
Antonetti, D. A., Silva, P. S. & Stitt, A. W. Current understanding of the molecular and cellular pathology of diabetic retinopathy. Nat. Rev. Endocrinol. 17, 195–206 (2021).
Nakano, T. et al. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell 10, 771–785 (2012). One of the founding articles of the organoid field describing the formation of the complex architecture of the eye from pluripotent stem cells.
Zhong, X. et al. Generation of three-dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat. Commun. 5, 4047 (2014).
O’Hara-Wright, M. & Gonzalez-Cordero, A. Retinal organoids: a window into human retinal development. Development 147, dev189746 (2020).
Achberger, K. et al. Merging organoid and organ-on-a-chip technology to generate complex multi-layer tissue models in a human retina-on-a-chip platform. Elife 8, e46188 (2019).
Blondeau, B., Avril, I., Duchene, B. & Bréant, B. Endocrine pancreas development is altered in foetuses from rats previously showing intra-uterine growth retardation in response to malnutrition. Diabetologia 45, 394–401 (2002).
Reusens, B., Theys, N., Dumortier, O., Goosse, K. & Remacle, C. Maternal malnutrition programs the endocrine pancreas in progeny. Am. J. Clin. Nutr. 94, 1824S–1829S (2011).
Chamson-Reig, A., Thyssen, S. M., Arany, E. & Hill, D. J. Altered pancreatic morphology in the offspring of pregnant rats given reduced dietary protein is time and gender specific. J. Endocrinol. 191, 83–92 (2006).
Calzada, L. et al. Maternal protein restriction during gestation impairs female offspring pancreas development in the rat. Nutr. Res. 36, 855–862 (2016).
King, R. et al. Offspring of mice exposed to a low-protein diet in utero demonstrate changes in mTOR signaling in pancreatic islets of Langerhans, associated with altered glucagon and insulin expression and a lower β-cell mass. Nutrients 11, 605 (2019).
Alejandro, E. U. et al. Maternal low-protein diet on the last week of pregnancy contributes to insulin resistance and β-cell dysfunction in the mouse offspring. Am. J. Physiol. Integr. Comp. Physiol. 319, R485–R496 (2020).
Han, J., Xu, J., Epstein, P. N. & Liu, Y. Q. Long-term effect of maternal obesity on pancreatic beta cells of offspring: reduced beta cell adaptation to high glucose and high-fat diet challenges in adult female mouse offspring. Diabetologia 48, 1810–1818 (2005).
Bringhenti, I. et al. Maternal obesity during the preconception and early life periods alters pancreatic development in early and adult life in male mouse offspring. PLoS ONE 8, e55711 (2013).
Ohta, T. et al. Maternal high-fat diet promotes onset of diabetes in rat offspring. Anim. Sci. J. 88, 149–155 (2017).
Nicholas, L. M. et al. Exposure to maternal obesity programs sex differences in pancreatic islets of the offspring in mice. Diabetologia 63, 324–337 (2020).
Bodin, J. et al. Transmaternal bisphenol A exposure accelerates diabetes type 1 development in NOD mice. Toxicol. Sci. 137, 311–323 (2014).
Alonso-Magdalena, P., García-Arévalo, M., Quesada, I. & Nadal, Á. Bisphenol-A treatment during pregnancy in mice: a new window of susceptibility for the development of diabetes in mothers later in life. Endocrinology 156, 1659–1670 (2015).
Whitehead, R., Guan, H., Arany, E., Cernea, M. & Yang, K. Prenatal exposure to bisphenol A alters mouse fetal pancreatic morphology and islet composition. Horm. Mol. Biol. Clin. Investig. 25, 171–179 (2016).
García-Arévalo, M. et al. Maternal exposure to bisphenol-A during pregnancy increases pancreatic β-cell growth during early life in male mice offspring. Endocrinology 157, 4158–4171 (2016).
La Merrill, M. A. et al. Consensus on the key characteristics of endocrine-disrupting chemicals as a basis for hazard identification. Nat. Rev. Endocrinol. 16, 45–57 (2020).
Nadal, A. et al. Bisphenol-A alters pancreatic B-cell proliferation and mass in an estrogen receptor beta-dependent manner [abstract]. J. Endocr. Soc. 4 (Suppl. 1), SAT-715 (2020).
Malmqvist, E. et al. Maternal exposure to air pollution and type 1 diabetes – accounting for genetic factors. Environ. Res. 140, 268–274 (2015).
Gillies, R. et al. Maternal exposure to Δ9-tetrahydrocannabinol impairs female offspring glucose homeostasis and endocrine pancreatic development in the rat. Reprod. Toxicol. 94, 84–91 (2020).
Moullé, V. S. & Parnet, P. Effects of nutrient intake during pregnancy and lactation on the endocrine pancreas of the offspring. Nutrients 11, 2708 (2019).
Lahti-Pulkkinen, M. et al. Consequences of being overweight or obese during pregnancy on diabetes in the offspring: a record linkage study in Aberdeen, Scotland. Diabetologia 62, 1412–1419 (2019).
Perng, W., Oken, E. & Dabelea, D. Developmental overnutrition and obesity and type 2 diabetes in offspring. Diabetologia 62, 1779–1788 (2019).
Sandovici, I. et al. Maternal diet and aging alter the epigenetic control of a promoter-enhancer interaction at the Hnf4a gene in rat pancreatic islets. Proc. Natl Acad. Sci. USA 108, 5449–5454 (2011).
Vaiserman, A. & Lushchak, O. Prenatal malnutrition-induced epigenetic dysregulation as a risk factor for type 2 diabetes. Int. J. Genomics 2019, 3821409 (2019).
Kadayifci, F. Z. et al. Early-life programming of type 2 diabetes mellitus: understanding the association between epigenetics/genetics and environmental factors. Curr. Genomics 20, 453–463 (2019).
Beyaz, S. et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature 531, 53–58 (2016).
Shang, L. et al. β-Cell dysfunction due to increased ER stress in a stem cell model of Wolfram syndrome. Diabetes 63, 923–933 (2014).
Montaser, H. et al. Loss of MANF causes childhood-onset syndromic diabetes due to increased endoplasmic reticulum stress. Diabetes 70, 1006–1018 (2021).
Maxwell, K. G. et al. Gene-edited human stem cell-derived β cells from a patient with monogenic diabetes reverse preexisting diabetes in mice. Sci. Transl. Med. 12, eaax9106 (2020).
Shik Mun, K. et al. Patient-derived pancreas-on-a-chip to model cystic fibrosis-related disorders. Nat. Commun. 10, 3124 (2019).
Halban, P. A. et al. The possible importance of contact between pancreatic islet cells for the control of insulin release. Endocrinology 111, 86–94 (1982). This historical article shows the benefits of endocrine cell re-aggregation for improving cellular function.
Tsonkova, V. G. et al. The EndoC-βH1 cell line is a valid model of human beta cells and applicable for screenings to identify novel drug target candidates. Mol. Metab. 8, 144–157 (2018).
Reissaus, C. A. & Piston, D. W. Reestablishment of glucose inhibition of glucagon secretion in small pseudoislets. Diabetes 66, 960–969 (2017).
Broutier, L. et al. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 11, 1724–1743 (2016).
Harata, M. et al. Delivery of shRNA via lentivirus in human pseudoislets provides a model to test dynamic regulation of insulin secretion and gene function in human islets. Physiol. Rep. 6, e13907 (2018).
Liu, S. et al. Lentiviral mediated gene silencing in human pseudoislet prepared in low attachment plates. J. Vis. Exp. 2019, 139–148 (2019).
Bevacqua, R. J. et al. SIX2 and SIX3 coordinately regulate functional maturity and fate of human pancreatic β cells. Genes Dev. 35, 234–249 (2021).
Author information
Authors and Affiliations
Contributions
A.G.-B. and B.S.B.-T. researched data for the article, contributed substantially to discussion of content, wrote the article and reviewed/edited the article before submission. S.Y. researched data for the article, contributed substantially to discussion of content and wrote the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Endocrinology thanks Mostafa Bakhti, Françoise Carlotti and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Beydag-Tasöz, B.S., Yennek, S. & Grapin-Botton, A. Towards a better understanding of diabetes mellitus using organoid models. Nat Rev Endocrinol 19, 232–248 (2023). https://doi.org/10.1038/s41574-022-00797-x
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41574-022-00797-x
This article is cited by
-
Analysis of high glucose injury using human induced pluripotent stem cell-derived kidney organoids
BMC Nephrology (2026)
-
Models of hyperglycaemia in diabetes mellitus and its complications
Nature Reviews Endocrinology (2026)
-
Advances in liver and pancreas organoids: how far we have come and where we go next
Nature Reviews Gastroenterology & Hepatology (2026)
-
Advances and continuing challenges in differentiation of stem cells to human kidney tissue
Nature Reviews Nephrology (2026)
-
Scaffold-free endocrine tissue engineering: role of islet organization and implications in type 1 diabetes
BMC Endocrine Disorders (2025)