Abstract
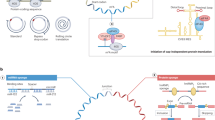
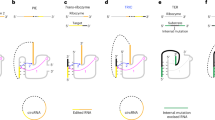
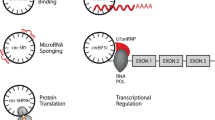
Over the past decade, research into circular RNA (circRNA) has increased rapidly, and over the past few years, circRNA has emerged as a promising therapeutic platform. The regulatory functions of circRNAs, including their roles in templating protein translation and regulating protein and RNA functions, as well as their unique characteristics, such as increased stability and a favourable immunological profile compared with mRNAs, make them attractive candidates for RNA-based therapies. Here, we describe the properties of circRNAs, their therapeutic potential and technologies for their synthesis. We also discuss the prospects and challenges to be overcome to unlock the full potential of circRNAs as drugs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Hogan, M. J. & Pardi, N. mRNA vaccines in the COVID-19 pandemic and beyond. Annu. Rev. Med. 73, 17–39 (2022).
Zhang, Y. et al. The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624 (2016).
Enuka, Y. et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383 (2016).
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J. & Kleinschmidt, A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA 73, 3852–3856 (1976).
Nigro, J. M. et al. Scrambled exons. Cell 64, 607–613 (1991).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691 (2019).
Liu, C. X. & Chen, L. L. Circular RNAs: characterization, cellular roles, and applications. Cell 185, 2016–2034 (2022).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629 (2018). This study optimizes PIE circRNA synthesis and uses exogenous circRNAs for robust and stable protein expression in eukaryotic cells, showing that circRNA is a promising alternative to linear mRNA.
Piwecka, M. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422 (2011).
Lebreton, A., Tomecki, R., Dziembowski, A. & Séraphin, B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature 456, 993–996 (2008).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e21 (2019).
Unti, M. J. & Jaffrey, S. R. Highly efficient cellular expression of circular mRNA enables prolonged protein expression. Cell Chem. Biol. 31, 163–176.e5 (2024).
Tai, J. & Chen, Y. G. Differences in the immunogenicity of engineered circular RNAs. J. Mol. Cell Biol. 15, mjad002 (2023). A review article providing a detailed discussion of the immunogenicity of IVT-derived circRNA.
Chen, Y. G. et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell 67, 228–238.e5 (2017).
Chen, Y. G. et al. N6-methyladenosine modification controls circular RNA immunity. Mol. Cell 76, 96–109.e9 (2019).
Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell 74, 508–520.e4 (2019). This study reports exogenous circRNA delivery and translation in vivo, showing that circRNA translation persists for longer than that of both unmodified and uridine-modified linear mRNAs.
Liu, C. X. et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell 82, 420–434.e6 (2022).
Guo, S. K. et al. Therapeutic application of circular RNA aptamers in a mouse model of psoriasis. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02204-4 (2024). This study shows that lipid nanoparticle-delivered double-stranded circRNAs can inhibit PKR activity, reduce inflammatory signals and alleviate symptoms of psoriasis, highlighting their potential as a therapeutic strategy.
Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 41, 262–272 (2023). This study optimizes IVT circRNA design to markedly enhance protein production by improving key elements such as vector topology, untranslated regions and translation initiation factors.
Chen, H. et al. Chemical and topological design of multicapped mRNA and capped circular RNA to augment translation. Nat. Biotechnol. https://doi.org/10.1038/s41587-024-02393-y (2024).
Xie, J. et al. Circular RNA: a promising new star of vaccine. J. Transl. Intern. Med. 11, 372–381 (2023).
Niu, D., Wu, Y. & Lian, J. Circular RNA vaccine in disease prevention and treatment. Signal Transduct. Target. Ther. 8, 341 (2023).
Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744.e16 (2022). This study shows the successful use of IVT-derived circRNA for vaccination against SARS-CoV-2 in vivo.
Seephetdee, C. et al. A circular mRNA vaccine prototype producing VFLIP-X spike confers a broad neutralization of SARS-CoV-2 variants by mouse sera. Antivir. Res. 204, 105370 (2022).
Wan, J. et al. Circular RNA vaccines with long-term lymph node-targeting delivery stability after lyophilization induce potent and persistent immune responses. mBio 15, e0177523 (2024).
Li, H. et al. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics 12, 6422–6436 (2022).
Yang, J. et al. Intratumoral delivered novel circular mRNA encoding cytokines for immune modulation and cancer therapy. Mol. Ther. Nucleic Acids 30, 184–197 (2022).
Feng, Z. et al. An in vitro-transcribed circular RNA targets the mitochondrial inner membrane cardiolipin to ablate EIF4G2+/PTBP1+ pan-adenocarcinoma. Nat. Cancer 5, 30–46 (2024).
Garber, K. Orna Therapeutics: circular logic. Nat. Biotechnol. https://doi.org/10.1038/d41587-022-00005-1 (2022).
Wang, Y. et al. Synergically enhanced anti-tumor immunity of in vivo CAR by circRNA vaccine boosting. Preprint at bioRxiv https://doi.org/10.1101/2024.07.05.600312 (2024).
Shen, L. et al. Circular mRNA-based TCR-T offers a safe and effective therapeutic strategy for treatment of cytomegalovirus infection. Mol. Ther. 32, 168–184 (2024).
Mullard, A. In vivo CAR T cells move into clinical trials. Nat. Rev. Drug Discov. 23, 727–730 (2024).
Hollensen, A. K. et al. Enhanced tailored microRNA sponge activity of RNA Pol II-transcribed TuD hairpins relative to ectopically expressed ciRS7-derived circRNAs. Mol. Ther. Nucleic Acids 13, 365–375 (2018).
Lavenniah, A. et al. Engineered circular RNA sponges act as miRNA inhibitors to attenuate pressure overload-induced cardiac hypertrophy. Mol. Ther. 28, 1506–1517 (2020).
Breuer, J. & Rossbach, O. Production and purification of artificial circular RNA sponges for application in molecular biology and medicine. Methods Protoc. 3, 42 (2020).
Jost, I. et al. Functional sequestration of microRNA-122 from Hepatitis C Virus by circular RNA sponges. RNA Biol. 15, 1032–1039 (2018).
Müller, S. et al. Synthetic circular miR-21 RNA decoys enhance tumor suppressor expression and impair tumor growth in mice. NAR Cancer 2, zcaa014 (2020).
Bayat, H., Pourgholami, M. H., Rahmani, S., Pournajaf, S. & Mowla, S. J. Synthetic miR-21 decoy circularized by tRNA splicing mechanism inhibited tumorigenesis in glioblastoma. Mol. Ther. Nucleic Acids 32, 432–444 (2023).
Liu, X. et al. Synthetic circular RNA functions as a miR-21 sponge to suppress gastric carcinoma cell proliferation. Mol. Ther. Nucleic Acids 13, 312–321 (2018).
Ren, S. et al. Efficient modulation of exon skipping via antisense circular RNAs. Research 6, 0045 (2023).
Pfafenrot, C. et al. Inhibition of SARS-CoV-2 coronavirus proliferation by designer antisense-circRNAs. Nucleic Acids Res. 49, 12502–12516 (2021).
Schreiner, S., Didio, A., Hung, L. H. & Bindereif, A. Design and application of circular RNAs with protein-sponge function. Nucleic Acids Res. 48, 12326–12335 (2020).
Siebring-van Olst, E. et al. A genome-wide siRNA screen for regulators of tumor suppressor p53 activity in human non-small cell lung cancer cells identifies components of the RNA splicing machinery as targets for anticancer treatment. Mol. Oncol. 11, 534–551 (2017).
Fei, T. et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl Acad. Sci. USA 114, E5207–E5215 (2017).
Zhou, X. et al. Abrogation of HnRNP L enhances anti-PD-1 therapy efficacy. Acta Pharm. Sin. B 12, 692–707 (2022).
Feng, X. et al. Circular RNA aptamers ameliorate AD-relevant phenotypes by targeting PKR. Preprint at bioRxiv https://doi.org/10.1101/2024.03.27.583257 (2024).
Umekage, S. & Kikuchi, Y. In vitro and in vivo production and purification of circular RNA aptamer. J. Biotechnol. 139, 265–272 (2009).
Litke, J. L. & Jaffrey, S. R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 37, 667–675 (2019). This study introduces the Tornado system, which facilitates rapid RNA circularization, resulting in highly stable and efficiently expressed circRNA aptamers.
Békés, M., Langley, D. R. & Crews, C. M. PROTAC targeted protein degraders: the past is prologue. Nat. Rev. Drug Discov. 21, 181–200 (2022).
Yang, J. et al. Circular mRNA encoded PROTAC (RiboPROTAC) as a new platform for the degradation of intracellular therapeutic targets. Preprint at bioRxiv https://doi.org/10.1101/2022.04.22.489232 (2022).
Liu, L. et al. Circular guide RNA for improved stability and CRISPR-Cas9 editing efficiency. ACS Synth. Biol. 12, 350–359 (2023).
Zhang, X. et al. Engineered circular guide RNAs boost CRISPR/Cas12a- and CRISPR/Cas13d-based DNA and RNA editing. Genome Biol. 24, 145 (2023).
Liu, B. et al. A split prime editor with untethered reverse transcriptase and circular RNA template. Nat. Biotechnol. 40, 1388–1393 (2022).
Liang, R. et al. Prime editing using CRISPR-Cas12a and circular RNAs in human cells. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02095-x (2024).
Katrekar, D. et al. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol. 40, 938–945 (2022).
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
Obi, P. & Chen, Y. G. The design and synthesis of circular RNAs. Methods 196, 85–103 (2021).
Petkovic, S. & Müller, S. RNA circularization strategies in vivo and in vitro. Nucleic Acids Res. 43, 2454–2465 (2015).
Bullard, D. R. & Bowater, R. P. Direct comparison of nick-joining activity of the nucleic acid ligases from bacteriophage T4. Biochem. J. 398, 135–144 (2006).
Moore, M. J. & Sharp, P. A. Site-specific modification of pre-mRNA: the 2’-hydroxyl groups at the splice sites. Science 256, 992–997 (1992).
Nandakumar, J., Ho, C. K., Lima, C. D. & Shuman, S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 279, 31337–31347 (2004).
Silber, R., Malathi, V. G. & Hurwitz, J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl Acad. Sci. USA 69, 3009–3013 (1972).
Cech, T. R. Self-splicing of group I introns. Annu. Rev. Biochem. 59, 543–568 (1990).
Ford, E. & Ares, M. Jr. Synthesis of circular RNA in bacteria and yeast using RNA cyclase ribozymes derived from a group I intron of phage T4. Proc. Natl Acad. Sci. USA 91, 3117–3121 (1994).
Puttaraju, M. & Been, M. D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 20, 5357–5364 (1992).
Cui, J. et al. A precise and efficient circular RNA synthesis system based on a ribozyme derived from Tetrahymena thermophila. Nucleic Acids Res. 51, e78 (2023).
Lee, K. H. et al. Efficient circular RNA engineering by end-to-end self-targeting and splicing reaction using Tetrahymena group I intron ribozyme. Mol. Ther. Nucleic Acids 33, 587–598 (2023).
Rausch, J. W. et al. Characterizing and circumventing sequence restrictions for synthesis of circular RNA in vitro. Nucleic Acids Res. 49, e35 (2021).
Pyle, A. M. Group II intron self-splicing. Annu. Rev. Biophys. 45, 183–205 (2016).
Jarrell, K. A. Inverse splicing of a group II intron. Proc. Natl Acad. Sci. USA 90, 8624–8627 (1993).
Chuyun, C. et al. A flexible, efficient, and scalable platform to produce circular RNAs as new therapeutics. Preprint at bioRxiv https://doi.org/10.1101/2022.05.31.494115 (2022).
Tong, M. et al. Robust genome and cell engineering via in vitro and in situ circularized RNAs. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-024-01245-z (2024).
Kim, Y. S. et al. The RNA ligation method using modified splint DNAs significantly improves the efficiency of circular RNA synthesis. Anim. Cell Syst. 27, 208–218 (2023).
Kaufmann, G., Klein, T. & Littauer, U. Z. T4 RNA ligase: substrate chain length requirements. FEBS Lett. 46, 271–275 (1974).
Sugino, A., Snoper, T. J. & Cozzarelli, N. R. Bacteriophage T4 RNA ligase. Reaction intermediates and interaction of substrates. J. Biol. Chem. 252, 1732–1738 (1977).
Ho, C. K. & Shuman, S. Bacteriophage T4 RNA ligase 2 (gp24.1) exemplifies a family of RNA ligases found in all phylogenetic domains. Proc. Natl Acad. Sci. USA 99, 12709–12714 (2002).
Suzuki, H. et al. Characterization of RNase R-digested cellular RNA source that consists of lariat and circular RNAs from pre-mRNA splicing. Nucleic Acids Res. 34, e63 (2006).
Xiao, M. S. & Wilusz, J. E. An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3′ ends. Nucleic Acids Res. 47, 8755–8769 (2019).
Panda, A. C. et al. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs. Nucleic Acids Res. 45, e116 (2017).
Abe, B. T., Wesselhoeft, R. A., Chen, R., Anderson, D. G. & Chang, H. Y. Circular RNA migration in agarose gel electrophoresis. Mol. Cell 82, 1768–1777.e3 (2022).
Chen, H. et al. Preferential production of RNA rings by T4 RNA ligase 2 without any splint through rational design of precursor strand. Nucleic Acids Res. 48, e54 (2020).
Carmona, E. M. Circular RNA: Design Criteria for Optimal Therapeutical Utility. PhD thesis, Harvard Univ. (2019).
Kameda, S., Ohno, H. & Saito, H. Synthetic circular RNA switches and circuits that control protein expression in mammalian cells. Nucleic Acids Res. 51, e24 (2023).
Capel, B. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030 (1993).
Dubin, R. A., Kazmi, M. A. & Ostrer, H. Inverted repeats are necessary for circularization of the mouse testis Sry transcript. Gene 167, 245–248 (1995).
Pasman, Z., Been, M. D. & Garcia-Blanco, M. A. Exon circularization in mammalian nuclear extracts. RNA 2, 603–610 (1996).
Zaphiropoulos, P. G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: correlation with exon skipping. Proc. Natl Acad. Sci. USA 93, 6536–6541 (1996).
Zaphiropoulos, P. G. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol. Cell. Biol. 17, 2985–2993 (1997).
Surono, A. et al. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum. Mol. Genet. 8, 493–500 (1999).
Li, X. F. & Lytton, J. A circularized sodium-calcium exchanger exon 2 transcript. J. Biol. Chem. 274, 8153–8160 (1999).
Gualandi, F. et al. Multiple exon skipping and RNA circularisation contribute to the severe phenotypic expression of exon 5 dystrophin deletion. J. Med. Genet. 40, e100 (2003).
Burd, C. E. et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6, e1001233 (2010).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). A landmark study detailing the biological function of the endogenous circRNA ciRS-7 as a miRNA sponge.
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013). A landmark study detailing the prevalence of circRNAs across eukaryotes.
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7, e30733 (2012).
Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell 159, 134–147 (2014).
Ivanov, A. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177 (2015).
Aktaş, T. et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119 (2017).
Liang, D. & Wilusz, J. E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28, 2233–2247 (2014).
Meganck, R. M. et al. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 23, 821–834 (2021). This study demonstrates the use of AAV for vector-based expression of protein-coding circRNAs, using inverted repeats to promote efficient backsplicing.
Wang, Y. & Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 21, 172–179 (2015). This study explores the use of plasmids for vector-based expression of protein-coding circRNAs, using inverted repeats to enhance backsplicing efficiency.
Hansen, T. B. in Circular RNAs. Methods in Molecular Biology, Vol. 1724 (eds Dieterich, C. & Papantonis, A.) 143–157 (Humana Press, 2018).
Stagsted, L. V. W., O’Leary, E. T., Ebbesen, K. K. & Hansen, T. B. The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. eLife 10, e63088 (2021).
Ho-Xuan, H. et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 48, 10368–10382 (2020).
Chu, J., Robert, F. & Pelletier, J. Trans-spliced mRNA products produced from circRNA expression vectors. RNA 27, 676–682 (2021).
Yang, W., Du, W. W., Li, X., Yee, A. J. & Yang, B. B. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene 35, 3919–3931 (2016).
Paludan, S. R. & Bowie, A. G. Immune sensing of DNA. Immunity 38, 870–880 (2013).
Young, J. L., Benoit, J. N. & Dean, D. A. Effect of a DNA nuclear targeting sequence on gene transfer and expression of plasmids in the intact vasculature. Gene Ther. 10, 1465–1470 (2003).
Le Guen, Y. T. et al. DNA nuclear targeting sequences for enhanced non-viral gene transfer: an in vitro and in vivo study. Mol. Ther. Nucleic Acids 24, 477–486 (2021).
Brown, D. W. et al. Safe and effective in vivo delivery of DNA and RNA using proteolipid vehicles. Cell 187, 5357–5375.e24 (2024).
Ertl, H. C. J. Immunogenicity and toxicity of AAV gene therapy. Front. Immunol. 13, 975803 (2022).
Meganck, R. M. et al. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol. Ther. Nucleic Acids 13, 89–98 (2018).
Lui, H. et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics 11, 1396–1411 (2021).
Mecozzi, N. et al. Genetic tools for the stable overexpression of circular RNAs. RNA Biol. 19, 353–363 (2022).
Nemeth, K., Bayraktar, R., Ferracin, M. & Calin, G. A. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat. Rev. Genet. 25, 211–232 (2024).
Zhao, R. J., Zhang, W. Y. & Fan, X. X. Circular RNAs: potential biomarkers and therapeutic targets for autoimmune diseases. Heliyon 10, e23694 (2024).
D’Anca, M., Buccellato, F. R., Fenoglio, C. & Galimberti, D. Circular RNAs: emblematic players of neurogenesis and neurodegeneration. Int. J. Mol. Sci. 23, 4134 (2022).
van Zonneveld, A. J., Kölling, M., Bijkerk, R. & Lorenzen, J. M. Circular RNAs in kidney disease and cancer. Nat. Rev. Nephrol. 17, 814–826 (2021).
Zhang, C. et al. Rapid development of targeting circRNAs in cardiovascular diseases. Mol. Ther. Nucleic Acids 21, 568–576 (2020).
Dehghanbanadaki, H. et al. Diagnostic accuracy of circular RNA for diabetes mellitus: a systematic review and diagnostic meta-analysis. BMC Med. Genomics 16, 48 (2023).
Kristensen, L. S., Hansen, T. B., Venø, M. T. & Kjems, J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene 37, 555–565 (2018).
Kristensen, L. S., Jakobsen, T., Hager, H. & Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 19, 188–206 (2022).
Li, Y. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015).
Fu, M. et al. Extracellular vesicles containing circMYBL1 induce CD44 in adenoid cystic carcinoma cells and pulmonary endothelial cells to promote lung metastasis. Cancer Res. 84, 2484–2500 (2024).
Vo, J. N. et al. The landscape of circular RNA in cancer. Cell 176, 869–881.e13 (2019).
Salachan, P. V. et al. Microbiota of the prostate tumor environment investigated by whole-transcriptome profiling. Genome Med. 14, 9 (2022).
Wen, N. et al. Cholangiocarcinoma combined with biliary obstruction: an exosomal circRNA signature for diagnosis and early recurrence monitoring. Signal Transduct. Target. Ther. 9, 107 (2024).
Dahl, M. et al. Expression patterns and prognostic potential of circular RNAs in mantle cell lymphoma: a study of younger patients from the MCL2 and MCL3 clinical trials. Leukemia 36, 177–188 (2022).
Salim, R. et al. Exploring new prognostic biomarkers in Mantle Cell Lymphoma: a comparison of the circSCORE and the MCL35 score. Leuk. Lymphoma 64, 1414–1423 (2023).
Papatsirou, M. et al. Exploring the molecular biomarker utility of circCCT3 in multiple myeloma: a favorable prognostic indicator, particularly for R-ISS II patients. HemaSphere 8, e34 (2024).
Bachmayr-Heyda, A. et al. Correlation of circular RNA abundance with proliferation — exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 5, 8057 (2015).
García-Rodríguez, J. L. et al. Spatial profiling of circular RNAs in cancer reveals high expression in muscle and stromal cells. Cancer Res. 83, 3340–3353 (2023).
Dong, Y. et al. Identification of circRNA signature associated with tumor immune infiltration to predict therapeutic efficacy of immunotherapy. Nat. Commun. 14, 2540 (2023).
Moldovan, L. I. et al. High-throughput RNA sequencing from paired lesional- and non-lesional skin reveals major alterations in the psoriasis circRNAome. BMC Med. Genomics 12, 174 (2019).
Moldovan, L. I. et al. Characterization of circular RNA transcriptomes in psoriasis and atopic dermatitis reveals disease-specific expression profiles. Exp. Dermatol. 30, 1187–1196 (2021).
Holdt, L. M. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7, 12429 (2016).
Fasolo, F. et al. The circular RNA Ataxia Telangiectasia Mutated regulates oxidative stress in smooth muscle cells in expanding abdominal aortic aneurysms. Mol. Ther. Nucleic Acids 33, 848–865 (2023).
Dong, X. et al. Circular RNAs in the human brain are tailored to neuron identity and neuropsychiatric disease. Nat. Commun. 14, 5327 (2023).
Hampel, H. et al. Designing the next-generation clinical care pathway for Alzheimer’s disease. Nat. Aging 2, 692–703 (2022).
Jiang, B. et al. Circulating exosomal hsa_circRNA_0039480 is highly expressed in gestational diabetes mellitus and may be served as a biomarker for early diagnosis of GDM. J. Transl. Med. 20, 5 (2022).
Xu, S. et al. Tumor-tailored ionizable lipid nanoparticles facilitate IL-12 circular RNA delivery for enhanced lung cancer immunotherapy. Adv. Mater. 36, e2400307 (2024).
Chen, C. Y. & Sarnow, P. Initiation of protein synthesis by the eukaryotic translational apparatus on circular RNAs. Science 268, 415–417 (1995).
Abe, N. et al. Rolling circle amplification in a prokaryotic translation system using small circular RNA. Angew. Chem. Int. Ed. 52, 7004–7008 (2013).
Bohjanen, P. R., Colvin, R. A., Puttaraju, M., Been, M. D. & Garcia-Blanco, M. A. A small circular TAR RNA decoy specifically inhibits Tat-activated HIV-1 transcription. Nucleic Acids Res. 24, 3733–3738 (1996).
Kajimoto, S. et al. Enzymatic conjugation of modified RNA fragments by ancestral RNA ligase AncT4_2. Appl. Environ. Microbiol. 88, e01679-22 (2022).
Kestemont, D. et al. XNA ligation using T4 DNA ligase in crowding conditions. Chem. Commun. 54, 6408–6411 (2018).
Goffin, C., Bailly, V. & Verly, W. G. Nicks 3′ or 5′ to AP sites or to mispaired bases, and one-nucleotide gaps can be sealed by T4 DNA ligase. Nucleic Acids Res. 15, 8755–8771 (1987).
Turunen, J. J. et al. in Handbook of RNA Biochemistry (eds Hartmann, R. K. et al.) 45–88 (Wiley, 2014).
Abe, N., Kodama, A. & Abe, H. in Circular RNAs. Methods in Molecular Biology, Vol. 1724 (eds Dieterich, C. & Papantonis, A.) 181–192 (Humana Press, 2018).
Abe, N. et al. Rolling circle translation of circular RNA in living human cells. Sci. Rep. 5, 16435 (2015).
Rigden, J. E. & Rezaian, M. A. In vitro synthesis of an infectious viroid: analysis of the infectivity of monomeric linear CEV. Virology 186, 201–206 (1992).
Bain, J. D. & Switzer, C. Regioselective ligation of oligoribonucleotides using DNA splints. Nucleic Acids Res. 20, 4372 (1992).
Beadudry, D. & Perreault, J.-P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 23, 3064–3066 (1995).
Wang, L. & Ruffner, D. E. Oligoribonucleotide circularization by ‘template-mediated’ ligation with T4 RNA ligase: synthesis of circular hammerhead ribozymes. Nucleic Acids Res. 26, 2502–2504 (1998).
Abe, N. et al. Synthesis, structure, and biological activity of dumbbell-shaped nanocircular RNAs for RNA interference. Bioconjugate Chem. 22, 2082–2092 (2011).
Breuer, J. et al. What goes around comes around: artificial circular RNAs bypass cellular antiviral responses. Mol. Ther. Nucleic Acids 28, 623–635 (2022).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 (2014).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Kristensen, L. S. et al. Spatial expression analyses of the putative oncogene ciRS-7 in cancer reshape the microRNA sponge theory. Nat. Commun. 11, 4551 (2020).
Dahl, M. et al. Enzyme-free digital counting of endogenous circular RNA molecules in B-cell malignancies. Lab. Invest. 98, 1657–1669 (2018).
Acknowledgements
This work was funded by a grant from the Danish National Research Foundation to the Centre for Cellular Signal Patterns (CellPAT) (grant DNRF 135) and by the Novo Nordisk Foundation (grant NNF23OC0081177). The authors thank M. Gockert for proofreading the manuscript and for valuable comments.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article. All authors contributed substantially to discussion of the content. All authors wrote the article. All authors reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
E.O’L. and T.B.H. are employees of Circio AB, which is developing vector-based circular RNA therapeutics. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
O’Leary, E., Jiang, Y., Kristensen, L.S. et al. The therapeutic potential of circular RNAs. Nat Rev Genet 26, 230–244 (2025). https://doi.org/10.1038/s41576-024-00806-x
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41576-024-00806-x
This article is cited by
-
Unlocking the power of non-coding RNAs: toward real-time cancer monitoring in precision oncology
Molecular Cancer (2026)
-
CircZBTB46 alleviates metabolic dysfunction–associated steatotic liver disease by targeting miRNA-326/FGF1 axis
Cell Death Discovery (2026)
-
The emerging role of epigenetic regulation in pain sensitization associated with headache disorders
The Journal of Headache and Pain (2025)
-
Functions and applications of enzymes in nucleic acid nanotechnology
Journal of Nanobiotechnology (2025)
-
Extracellular vesicle-mediated delivery of circp53 suppresses the progression of multiple cancers by activating the CypD/TRAP/HSP90 pathway
Experimental & Molecular Medicine (2025)