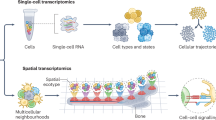
Abstract
Spatial transcriptomics is a powerful method for studying the spatial organization of cells, which is a critical feature in the development, function and evolution of multicellular life. However, sequencing-based spatial transcriptomics has not yet achieved cellular-level resolution, so advanced deconvolution methods are needed to infer cell-type contributions at each location in the data. Recent progress has led to diverse tools for cell-type deconvolution that are helping to describe tissue architectures in health and disease. In this Review, we describe the varied types of cell-type deconvolution methods for spatial transcriptomics, contrast their capabilities and summarize them in a web-based, interactive table to enable more efficient method selection.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lecuit, T. & Lenne, P.-F. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat. Rev. Mol. Cell Biol. 8, 633–644 (2007).
Zeller, R., López-Ríos, J. & Zuniga, A. Vertebrate limb bud development: moving towards integrative analysis of organogenesis. Nat. Rev. Genet. 10, 845–858 (2009).
Egeblad, M., Nakasone, E. S. & Werb, Z. Tumors as organs: complex tissues that interface with the entire organism. Dev. Cell 18, 884–901 (2010).
Takeichi, M. Dynamic contacts: rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15, 397–410 (2014).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Stark, R., Grzelak, M. & Hadfield, J. RNA sequencing: the teenage years. Nat. Rev. Genet. 20, 631–656 (2019).
Ståhl, P. L. et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science 353, 78–82 (2016).
Moses, L., Pachter, L. Museum of spatial transcriptomics. Nat Methods 19, 534–546 (2022). This Review introduces spatial transcriptomics technologies and its core concepts and tools for data analysis.
Tian, L., Chen, F. & Macosko, E. Z. The expanding vistas of spatial transcriptomics. Nat Biotechnol 41, 773–782 (2023).
Rodriques, S. G. et al. Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363, 1463–1467 (2019).
Liu, Y. et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell 183, 1665–1681.e18 (2020).
Merritt, C. R. et al. Multiplex digital spatial profiling of proteins and RNA in fixed tissue. Nat. Biotechnol. 38, 586–599 (2020).
Korsunsky, I. et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat. Methods 16, 1289–1296 (2019).
Welch, J. D. et al. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887.e17 (2019).
Chen, B., Khodadoust, M. S., Liu, C. L., Newman, A. M. & Alizadeh, A. A. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol. Biol. 1711, 243–259 (2018).
Avila Cobos, F., Alquicira-Hernandez, J., Powell, J. E., Mestdagh, P. & De Preter, K. Benchmarking of cell type deconvolution pipelines for transcriptomics data. Nat. Commun. 11, 5650 (2020).
Zhang, Y. et al. Deconvolution algorithms for inference of the cell-type composition of the spatial transcriptome. Comput. Struct. Biotechnol. J. 21, 176–184 (2023).
Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. 22, 627–644 (2021).
Garmire, L. X. et al. Challenges and perspectives in computational deconvolution of genomics data. Nat. Methods 21, 391–400 (2024).
Gaujoux, R. & Seoighe, C. CellMix: a comprehensive toolbox for gene expression deconvolution. Bioinformatics 29, 2211–2212 (2013).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12, 453–457 (2015).
Wang, X., Park, J., Susztak, K., Zhang, N. R. & Li, M. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 10, 380 (2019).
Tsoucas, D. et al. Accurate estimation of cell-type composition from gene expression data. Nat. Commun. 10, 2975 (2019).
Dong, R. & Yuan, G.-C. SpatialDWLS: accurate deconvolution of spatial transcriptomic data. Genome Biol. 22, 145 (2021).
Xun, Z. et al. Reconstruction of the tumor spatial microenvironment along the malignant–boundary–nonmalignant axis. Nat. Commun. 14, 933 (2023).
Yang, T. et al. AdRoit is an accurate and robust method to infer complex transcriptome composition. Commun. Biol. 4, 1–14 (2021).
Chen, Y., Ruan, F. & Wang, J.-P. NLSDeconv: an efficient cell-type deconvolution method for spatial transcriptomics data. Bioinformatics 41, btae747 (2024).
Zhou, Z., Zhong, Y., Zhang, Z. & Ren, X. Spatial transcriptomics deconvolution at single-cell resolution using Redeconve. Nat. Commun. 14, 7930 (2023).
Elosua-Bayes, M., Nieto, P., Mereu, E., Gut, I. & Heyn, H. SPOTlight: seeded NMF regression to deconvolute spatial transcriptomics spots with single-cell transcriptomes. Nucleic Acids Res. 49, e50 (2021). This study showcases one of the first deconvolution methods for spatial transcriptomics data, SPOTlight, which combines matrix factorization with non-negative least squares regression to perform cell-type deconvolution.
Ru, B., Huang, J., Zhang, Y., Aldape, K. & Jiang, P. Estimation of cell lineages in tumors from spatial transcriptomics data. Nat. Commun. 14, 568 (2023).
Wei, R. et al. Spatial charting of single-cell transcriptomes in tissues. Nat. Biotechnol. 40, 1190–1199 (2022).
Swain, A. K., Pandit, V., Sharma, J. & Yadav, P. SpatialPrompt: spatially aware scalable and accurate tool for spot deconvolution and domain identification in spatial transcriptomics. Commun. Biol. 7, 1–15 (2024).
Lu, Y., Chen, Q. M. & An, L. SPADE: spatial deconvolution for domain specific cell-type estimation. Commun. Biol. 7, 1–12 (2024).
Danaher, P. et al. Advances in mixed cell deconvolution enable quantification of cell types in spatial transcriptomic data. Nat. Commun. 13, 385 (2022).
Liu, Z., Ma, L., Wu, D. & Zhai, W. SONAR enables cell type deconvolution with spatially weighted Poisson-gamma model for spatial transcriptomics. Nat. Commun. 14, 4727 (2023).
Ospina, O. E. et al. Differential gene expression analysis of spatial transcriptomic experiments using spatial mixed models. Sci. Rep. 14, 10967 (2024).
Ma, Y. & Zhou, X. Spatially informed cell-type deconvolution for spatial transcriptomics. Nat. Biotechnol. 40, 1349–1359 (2022). This article presents CARD, a method that combines dimensionality reduction (NMF) with a spatial model to integrate the coordinates in deconvolution results.
Chidester, B., Zhou, T., Alam, S. & Ma, J. SPICEMIX enables integrative single-cell spatial modeling of cell identity. Nat. Genet. 55, 78–88 (2023).
Georgaka, S. et al. CellPie: a scalable spatial transcriptomics factor discovery method via joint non-negative matrix factorization. Nucleic Acids Res. 53, gkaf251 (2025).
Zhang, M., Parker, J., An, L., Liu, Y. & Sun, X. Flexible analysis of spatial transcriptomics data (FAST): a deconvolution approach. BMC Bioinformatics 26, 35 (2025).
Sompairac, N. et al. Independent component analysis for unraveling the complexity of cancer omics datasets. Int. J. Mol. Sci. 20, 4414 (2019).
Captier, N. et al. BIODICA: a computational environment for independent component analysis of omics data. Bioinformatics 38, 2963–2964 (2022).
Thuilliez, C. et al. CellsFromSpace: a fast, accurate, and reference-free tool to deconvolve and annotate spatially distributed omics data. Bioinform. Adv. 4, vbae081 (2024).
Zhang, C. et al. LANTSA: landmark-based transferable subspace analysis for single-cell and spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2022.03.13.484116 (2022).
Andersson, A. et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 3, 1–8 (2020).
Lopez, R. et al. DestVI identifies continuums of cell types in spatial transcriptomics data. Nat. Biotechnol. 40, 1360–1369 (2022).
Kleshchevnikov, V. et al. Cell2location maps fine-grained cell types in spatial transcriptomics. Nat. Biotechnol. 40, 661–671 (2022). This article introduces Cell2location, a Bayesian cell-type deconvolution method that accounts for technical sources of variation and borrows statistical strength across locations. Cell2location is a top performer in published benchmarks.
Geras, A. et al. Celloscope: a probabilistic model for marker-gene-driven cell type deconvolution in spatial transcriptomics data. Genome Biol. 24, 120 (2023).
Li, C., Chan, T.-F., Yang, C. & Lin, Z. stVAE deconvolves cell-type composition in large-scale cellular resolution spatial transcriptomics. Bioinformatics 39, btad642 (2023).
Chen, J. et al. Cell composition inference and identification of layer-specific spatial transcriptional profiles with POLARIS. Sci. Adv. 9, eadd9818 (2023).
Zhu, S. et al. STIE: single-cell level deconvolution, convolution, and clustering in in situ capturing-based spatial transcriptomics. Nat. Commun. 15, 7559 (2024).
He, S. et al. Starfysh integrates spatial transcriptomic and histologic data to reveal heterogeneous tumor–immune hubs. Nat Biotechnol 43, 223–235 (2025).
Cable, D. M. et al. Robust decomposition of cell type mixtures in spatial transcriptomics. Nat. Biotechnol. 40, 517–526 (2022). This study introduces RCTD, a probabilistic deconvolution method and one of the current best performers. RCTD uses Poisson distribution to model spatial transcriptomics data and leverages single-cell RNA-sequencing to perform deconvolution.
Zhang, H. et al. BayesTME: an end-to-end method for multiscale spatial transcriptional profiling of the tissue microenvironment. Cell Syst. 14, 605–619.e7 (2023).
Luo, T. et al. MAST-Decon: smooth cell-type deconvolution method for spatial transcriptomics data. Preprint at bioRxiv https://doi.org/10.1101/2024.05.10.593595 (2024).
Geras, A., Domżał, K. & Szczurek, E. Joint cell type identification in spatial transcriptomics and single-cell RNA sequencing data. Preprint at bioRxiv https://doi.org/10.1101/2023.05.29.542559 (2024).
Niyakan, S. et al. MUSTANG: multi-sample spatial transcriptomics data analysis with cross-sample transcriptional similarity guidance. PATTER 5, 100986 (2024).
Singh, R. et al. RETROFIT: reference-free deconvolution of cell-type mixtures in spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2023.06.07.544126 (2023).
Wan, X. et al. Integrating spatial and single-cell transcriptomics data using deep generative models with SpatialScope. Nat. Commun. 14, 7848 (2023).
Zubair, A. et al. Cell type identification in spatial transcriptomics data can be improved by leveraging cell-type-informative paired tissue images using a Bayesian probabilistic model. Nucleic Acids Res. 50, e80 (2022).
Miller, B. F., Huang, F., Atta, L., Sahoo, A. & Fan, J. Reference-free cell type deconvolution of multi-cellular pixel-resolution spatially resolved transcriptomics data. Nat. Commun. 13, 2339 (2022). This article presents the first reference-free method tailored for deconvolution of spatial transcriptomics data, STdeconvolve, which uses Bayesian topic modelling to perform deconvolution.
Sun, D., Liu, Z., Li, T., Wu, Q. & Wang, C. STRIDE: accurately decomposing and integrating spatial transcriptomics using single-cell RNA sequencing. Nucleic Acids Res. 50, e42 (2022).
Yang, C. X., Sin, D. D. & Ng, R. T. SMART: spatial transcriptomics deconvolution using marker-gene-assisted topic model. Genome Biol. 25, 304 (2024).
Blei, D. M., Ng, A. Y. & Jordan, M. I. Latent Dirichlet allocation. J. Mach. Learn. Res. 3, 993–1022 (2003).
Eshima, S., Imai, K. & Sasaki, T. Keyword assisted topic models. Am. J. Pol. Sci. 68, 730–750 (2024).
Moriel, N. et al. NovoSpaRc: flexible spatial reconstruction of single-cell gene expression with optimal transport. Nat. Protoc. 16, 4177–4200 (2021).
Yang, P. et al. Revealing spatial multimodal heterogeneity in tissues with SpaTrio. Cell Genomics 3, 100446 (2023).
Mages, S. et al. TACCO unifies annotation transfer and decomposition of cell identities for single-cell and spatial omics. Nat. Biotechnol. 41, 1465–1473 (2023).
Nguyen, N. D. et al. scDOT: optimal transport for mapping senescent cells in spatial transcriptomics. Genome Biol. 25, 288 (2024).
Rahimi, A., Vale-Silva, L. A., Fälth Savitski, M., Tanevski, J. & Saez-Rodriguez, J. DOT: a flexible multi-objective optimization framework for transferring features across single-cell and spatial omics. Nat. Commun. 15, 4994 (2024).
Vahid, M. R. et al. High-resolution alignment of single-cell and spatial transcriptomes with CytoSPACE. Nat. Biotechnol. 41, 1543–1548 (2023).
Cang, Z. & Nie, Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat. Commun. 11, 2084 (2020).
Liao, J. et al. De novo analysis of bulk RNA-seq data at spatially resolved single-cell resolution. Nat. Commun. 13, 6498 (2022).
Lund, J. B. et al. AntiSplodge: a neural-network-based RNA-profile deconvolution pipeline designed for spatial transcriptomics. NAR Genomics Bioinform. 4, lqac073 (2022).
Xu, H. et al. SPACEL: deep learning-based characterization of spatial transcriptome architectures. Nat. Commun. 14, 7603 (2023).
Biancalani, T. et al. Deep learning and alignment of spatially resolved single-cell transcriptomes with Tangram. Nat. Methods 18, 1352–1362 (2021). Tangram is a pioneering deep-learning-based deconvolution method that learns a mapping function to maximize the spatial correlation of genes shared by single-cell RNA-sequencing and spatial data. Tangram is one of the most benchmarked methods.
Maseda, F., Cang, Z. & Nie, Q. DEEPsc: a deep learning-based map connecting single-cell transcriptomics and spatial imaging data. Front. Genet. 12, 636743 (2021).
Charytonowicz, D., Brody, R. & Sebra, R. Interpretable and context-free deconvolution of multi-scale whole transcriptomic data with UniCell Deconvolve. Nat. Commun. 14, 1350 (2023).
Bae, S., Choi, H. & Lee, D. S. spSeudoMap: cell type mapping of spatial transcriptomics using unmatched single-cell RNA-seq data. Genome Med. 15, 19 (2023).
Zhong, Y., Zhang, J. & Ren, X. Spatial transcriptomics prediction from histology images at single-cell resolution using RedeHist. Preprint at bioRxiv https://doi.org/10.1101/2024.06.17.599464 (2024).
Bae, S. et al. CellDART: cell type inference by domain adaptation of single-cell and spatial transcriptomic data. Nucleic Acids Res. 50, e57 (2022).
Zhao, C. et al. Innovative super-resolution in spatial transcriptomics: a transformer model exploiting histology images and spatial gene expression. Brief. Bioinform. 25, bbae052 (2024).
Hao, M. et al. STEM enables mapping of single-cell and spatial transcriptomics data with transfer learning. Commun. Biol. 7, 1–15 (2024).
Mañanes, D. et al. SpatialDDLS: an R package to deconvolute spatial transcriptomics data using neural networks. Bioinformatics 40, btae072 (2024).
Chen, H. et al. Recovering single-cell expression profiles from spatial transcriptomics with scResolve. Cell Rep. Methods 4, 100864 (2024).
Zhang, T. et al. GTAD: a graph-based approach for cell spatial composition inference from integrated scRNA-seq and ST-seq data. Brief. Bioinform. 25, bbad469 (2024).
Zhan, Y. et al. LETSmix: a spatially informed and learning-based domain adaptation method for cell-type deconvolution in spatial transcriptomics. Genome Med. 17, 16 (2025).
Coleman, K., Hu, J., Schroeder, A., Lee, E. B. & Li, M. SpaDecon: cell-type deconvolution in spatial transcriptomics with semi-supervised learning. Commun. Biol. 6, 1–13 (2023).
Yin, W., Wan, Y. & Zhou, Y. SpatialcoGCN: deconvolution and spatial information-aware simulation of spatial transcriptomics data via deep graph co-embedding. Brief. Bioinform. 25, bbae130 (2024).
Long, Y. et al. Spatially informed clustering, integration, and deconvolution of spatial transcriptomics with GraphST. Nat. Commun. 14, 1155 (2023).
Song, Q. & Su, J. DSTG: deconvoluting spatial transcriptomics data through graph-based artificial intelligence. Brief. Bioinform. 22, bbaa414 (2021).
Li, H., Li, H., Zhou, J. & Gao, X. SD2: spatially resolved transcriptomics deconvolution through integration of dropout and spatial information. Bioinformatics 38, 4878–4884 (2022).
Ding, J. et al. SpatialCTD: a large-scale tumor microenvironment spatial transcriptomic dataset to evaluate cell type deconvolution for immuno-oncology. J. Comput. Biol. 31, 871–885 (2024).
Li, Y. & Luo, Y. STdGCN: spatial transcriptomic cell-type deconvolution using graph convolutional networks. Genome Biol. 25, 206 (2024).
Valous, N. A., Popp, F., Zörnig, I., Jäger, D. & Charoentong, P. Graph machine learning for integrated multi-omics analysis. Br. J. Cancer 131, 205–211 (2024).
Regev, A. et al. The Human Cell Atlas. eLife 6, e27041 (2017).
Ando, Y., Kwon, A. T.-J. & Shin, J. W. An era of single-cell genomics consortia. Exp. Mol. Med. 52, 1409–1418 (2020).
Rood, J. E., Maartens, A., Hupalowska, A., Teichmann, S. A. & Regev, A. Impact of the Human Cell Atlas on medicine. Nat. Med. 28, 2486–2496 (2022).
Vallania, F. et al. Leveraging heterogeneity across multiple datasets increases cell-mixture deconvolution accuracy and reduces biological and technical biases. Nat. Commun. 9, 4735 (2018).
Li, H. et al. A comprehensive benchmarking with practical guidelines for cellular deconvolution of spatial transcriptomics. Nat. Commun. 14, 1548 (2023). This benchmark study reports the performances of 18 deconvolution methods and highlights the technology effect challenges, the importance of good documentation or tutorials and provides practical guidelines.
Li, B. et al. Benchmarking spatial and single-cell transcriptomics integration methods for transcript distribution prediction and cell type deconvolution. Nat. Methods 19, 662–670 (2022). This benchmark study focuses on ‘integration’ of spatial transcriptomics and single-cell RNA-sequencing through transcript distribution prediction and cell-type deconvolution.
Hao, Y. et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 42, 293–304 (2024).
Qiu, P. Embracing the dropouts in single-cell RNA-seq analysis. Nat. Commun. 11, 1169 (2020).
Baysoy, A., Bai, Z., Satija, R. & Fan, R. The technological landscape and applications of single-cell multi-omics. Nat. Rev. Mol. Cell Biol. 24, 695–713 (2023).
Young, M. D. & Behjati, S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9, giaa151 (2020).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Sturm, G. et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics 35, i436–i445 (2019).
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. CellPose: a generalist algorithm for cellular segmentation. Nat. Methods 18, 100–106 (2021).
Berg, S. et al. ilastik: interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019).
Song, A. H. et al. Artificial intelligence for digital and computational pathology. Nat. Rev. Bioeng. 1, 930–949 (2023).
Ronneberger, O., Fischer, P. & Brox, T. U-net: convolutional networks for biomedical image segmentation. In Int. Conf. Medical Image Computing and Computer-Assisted Intervention 234–241 (Springer, 2015).
Jaume, G. et al. HEST-1k: a dataset for spatial transcriptomics and histology image analysis. In Adv. Neural Information Processing Systems (NeurIPS, 2024). This article introduces a collection of spatial transcriptomics profiles, bringing together data from different spatial transcriptomics technologies along with whole-slide images with extensive metadata. They use the dataset to benchmark foundation models in pathology and to perform biomarker exploration.
Zormpas, E., Queen, R., Comber, A. & Cockell, S. J. Mapping the transcriptome: realizing the full potential of spatial data analysis. Cell 186, 5677–5689 (2023).
Engelhard, V. H. et al. Immune cell infiltration and tertiary lymphoid structures as determinants of antitumor immunity. J. Immunol. 200, 432–442 (2018).
Haley, M. J. et al. Hypoxia coordinates the spatial landscape of myeloid cells within glioblastoma to affect survival. Sci. Adv. 10, eadj3301 (2024).
Grout, J. A. et al. Spatial positioning and matrix programs of cancer-associated fibroblasts promote T-cell exclusion in human lung tumors. Cancer Discov. 12, 2606–2625 (2022).
Chen, J. et al. A comprehensive comparison on cell-type composition inference for spatial transcriptomics data. Brief. Bioinform. 23, bbac245 (2022). This benchmark study reports the performances of 10 deconvolution methods and includes a missing cell-type experiment in which deconvolution methods are tested for attributing transcriptomics signal of missing cell type in the reference.
Yan, L. & Sun, X. Benchmarking and integration of methods for deconvoluting spatial transcriptomic data. Bioinformatics 39, btac805 (2023). This benchmark study reports the performances of 14 deconvolution methods and proposes a new ensemble learning-based deconvolution method.
10x Genomics. Visium Datasets of Mouse Brain https://www.10xgenomics.com/datasets?configure%5BhitsPerPage%5D=50&configure%5BmaxValuesPerFacet%5D=1000&query=mouse%20brain&refinementList%5Bplatform%5D%5B0%5D=Visium%20Spatial.
Maynard, K. R. et al. Transcriptome-scale spatial gene expression in the human dorsolateral prefrontal cortex. Nat. Neurosci. 24, 425–436 (2021).
Moncada, R. et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat. Biotechnol. 38, 333–342 (2020).
Croizer, H. et al. Deciphering the spatial landscape and plasticity of immunosuppressive fibroblasts in breast cancer. Nat. Commun. 15, 2806 (2024).
Cheng, A., Hu, G. & Li, W. V. Benchmarking cell-type clustering methods for spatially resolved transcriptomics data. Brief. Bioinformatics 24, bbac475 (2023).
Hu, Y. et al. Benchmarking clustering, alignment, and integration methods for spatial transcriptomics. Genome Biol. 25, 212 (2024).
Moses, L. et al. Voyager: exploratory single-cell genomics data analysis with geospatial statistics. Preprint at bioRxiv https://doi.org/10.1101/2023.07.20.549945 (2023).
Zhao, E. et al. Spatial transcriptomics at subspot resolution with BayesSpace. Nat. Biotechnol. 39, 1375–1384 (2021).
Bergenstråhle, L. et al. Super-resolved spatial transcriptomics by deep data fusion. Nat. Biotechnol. 40, 476–479 (2022).
Hu, J. et al. Deciphering tumor ecosystems at super resolution from spatial transcriptomics with TESLA. Cell Syst. 14, 404–417.e4 (2023).
Zhang, D. et al. Inferring super-resolution tissue architecture by integrating spatial transcriptomics with histology. Nat. Biotechnol. 42, 1372–1377 (2024).
Tanevski, J., Flores, R. O. R., Gabor, A., Schapiro, D. & Saez-Rodriguez, J. Explainable multiview framework for dissecting spatial relationships from highly multiplexed data. Genome Biol. 23, 97 (2022).
Armingol, E., Baghdassarian, H. M. & Lewis, N. E. The diversification of methods for studying cell–cell interactions and communication. Nat. Rev. Genet. 25, 381–400 (2024).
Dries, R. et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 22, 78 (2021).
Virshup, I. et al. The scverse project provides a computational ecosystem for single-cell omics data analysis. Nat. Biotechnol. 41, 604–606 (2023).
Kueckelhaus, J. et al. Inferring histology-associated gene expression gradients in spatial transcriptomic studies. Nat. Commun. 15, 7280 (2024).
Zeira, R., Land, M., Strzalkowski, A. & Raphael, B. J. Alignment and integration of spatial transcriptomics data. Nat. Methods 19, 567–575 (2022).
Liu, W. et al. Probabilistic embedding, clustering, and alignment for integrating spatial transcriptomics data with PRECAST. Nat. Commun. 14, 296 (2023).
Velten, B. et al. Identifying temporal and spatial patterns of variation from multimodal data using MEFISTO. Nat. Methods 19, 179–186 (2022).
Deng, Y. et al. Spatial-CUT&Tag: spatially resolved chromatin modification profiling at the cellular level. Science 375, 681–686 (2022).
Zhang, D. et al. Spatial epigenome–transcriptome co-profiling of mammalian tissues. Nature 616, 113–122 (2023).
Liu, Y. et al. High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409 (2023).
Nunes, J. B. et al. Integration of mass cytometry and mass spectrometry imaging for spatially resolved single-cell metabolic profiling. Nat. Methods 21, 1796–1800 (2024).
Saeys, Y., Van Gassen, S. & Lambrecht, B. N. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 16, 449–462 (2016).
Saez-Rodriguez, J. et al. Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat. Rev. Genet. 17, 470–486 (2016).
DREAM Challenges https://dreamchallenges.org/about-dream/ (2025).
Stickels, R. R. et al. Highly sensitive spatial transcriptomics at near-cellular resolution with slide-seqV2. Nat. Biotechnol. 39, 313–319 (2021).
Moffitt, J. R. & Zhuang, X. RNA imaging with multiplexed error-robust fluorescence in situ hybridization (MERFISH). Methods Enzymol. 572, 1–49 (2016).
Janesick, A. et al. High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nat. Commun. 14, 8353 (2023).
Fan, Z., Chen, R. & Chen, X. SpatialDB: a database for spatially resolved transcriptomes. Nucleic Acids Res. 48, D233–D237 (2020).
Fan, Z. et al. SPASCER: spatial transcriptomics annotation at single-cell resolution. Nucleic Acids Res. 51, D1138–D1149 (2023).
Zheng, Y., Chen, Y., Ding, X., Wong, K. H. & Cheung, E. Aquila: a spatial omics database and analysis platform. Nucleic Acids Res. 51, D827–D834 (2023).
Li, Y. et al. SOAR elucidates disease mechanisms and empowers drug discovery through spatial transcriptomics. Preprint at bioRxiv https://doi.org/10.1101/2022.04.17.488596 (2023).
Wang, G. et al. CROST: a comprehensive repository of spatial transcriptomics. Nucleic Acids Res. 52, D882–D890 (2024).
Xu, Z. et al. STOmicsDB: a comprehensive database for spatial transcriptomics data sharing, analysis and visualization. Nucleic Acids Res. 52, D1053–D1061 (2024).
Polański, K. et al. Bin2cell reconstructs cells from high resolution Visium HD data. Bioinformatics 40, btae546 (2024).
Greenacre, M. Compositional data analysis. Annu. Rev. Stat. Appl. 8, 271–299 (2021). This review summarizes the key concepts and mathematical transformations of compositional data analysis.
Aitchison, J. The statistical analysis of compositional data. J. R. Stat. Soc. Ser. B 44, 139–160 (1982).
Pearson, K. Mathematical contributions to the theory of evolution. On a form of spurious correlation which may arise when indices are used in the measurement of organs. Proc. R. Soc. Lond. 60, 489–498 (1897).
Acknowledgements
L.C.G.-B. is supported by a Contrat Doctoral Spécifique Normalien (2022–2025), awarded by the École Normale Supérieure — PSL University. The work of L.C.G.-B. and F.M.G.C. has been performed with financial support from ITMO Cancer of Aviesan on funds Cancer 2021 administered by Inserm (ATIP-avenir), the Fondation pour la Recherche Medicale (FRM) (code dossier FRM: AJE201905008656), La Ville de Paris (programme emergence(s)), the Cancéropôle Paris region (Emergence 2021), l’INCA (2022-1-PL BIO-02-ICR-1), and La ligue contre le cancer, comité de Paris (RS23/75-43, RS24/75-67). The work of L.G., T.W. and E.B. was funded in part by the French government under management of Agence Nationale de la Recherche as part of the programmes ‘Investissements d’avenir’ (reference no. ANR-19-P3IA-0001; PRAIRIE 3IA Institute) and France 2030 (reference no. ANR-24-EXCI-0005). The authors also thank L. Chadoutaud for meaningful discussions and feedback.
Author information
Authors and Affiliations
Contributions
L.C.G.-B. and L.G. researched the literature. All authors contributed substantially to discussion of the content. L.C.G.-B. and L.G. wrote the article. All authors reviewed and edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Genetics thanks Jean Fan; Yvan Saeys, who co-reviewed with Chananchida Sang-aram; and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Update form: https://forms.gle/Cv3byanhgVAJBerk6
Web-based summary table: https://cavallilab-curie.shinyapps.io/Review-Deconvolution-for-Spatial-Transcriptomics/
Supplementary information
Glossary
- Archetypal analysis
-
A statistical technique to identify and summarize patterns in the data by finding archetypes. In deconvolution, these archetypes represent idealized spot composed of a unique cell type. Each spot is then represented as a mixture of these archetypes.
- Attention networks
-
Neural networks equipped with ‘attention mechanisms’ that enable the network to focus on relevant parts of the input data. Attention networks have been used extensively in Natural Language Processing and Computer Vision.
- Compositional data
-
Quantitative descriptions of the parts of some whole, conveying relative information. Common compositional data are proportions and percentages, as encountered in most deconvolution outputs.
- Dampened weighted least squares
-
A classical weighted least-squares regression method with two properties: weights are constrained to be greater than zero and a dampening constant is introduced to prevent infinite weights resulting from low cell-type proportions.
- Digital pathology
-
A branch of pathology that focuses on digitizing and analysing microscopic tissue slides, such as haematoxylin and eosin-stained samples, using high-resolution scanners and advanced artificial intelligence tools.
- Dimensionality reduction
-
A mathematical technique to represent high-dimensional data in lower dimensions. In the context of deconvolution, dimension reduction aims to represent the gene expression data as cell-type contribution in each spot.
- Domain shifts
-
A phenomenon in machine learning that occurs when training and test data do not follow the same distribution, leading to potential performance degradation. For example, staining protocols might differ between institutes and thus may affect model performance during image analyses.
- Ensemble learning
-
A machine-learning technique consisting in combining multiple models to obtain better predictive performances than individual models.
- Foundational models
-
Large-scale artificial intelligence models trained on broad unlabelled datasets that can be adapted for various downstream tasks.
- Fully connected neural network
-
A neural network in which each neuron in one layer is connected to all neurons in the next layer. Fully connected neural networks are highly flexible, but computationally expensive owing to a large number of parameters. For this reason, neural networks are often only partly fully connected.
- Graph neural networks
-
A type of neural network designed to process data structured as graphs.
- Haematoxylin and eosin
-
(H&E). A gold standard staining used in histology, in which the haematoxylin stains cell nuclei in purplish blue and eosin stains the extracellular matrix and the cytoplasm in pink.
- Maximum a posteriori
-
A probabilistic estimation method that finds the most likely parameter values given the observed data and a prior distribution.
- Monte Carlo algorithms
-
A computational method that relies on random sampling to approximate probability distributions and estimate model parameters. In Bayesian inference, Monte Carlo algorithms — such as Markov chain Monte Carlo — are used to generate samples from complex posterior distributions when direct analytical solutions are intractable.
- Multilayer perceptron
-
A type of fully connected neural network with multiple hidden layers, typically used for classification and regression tasks.
- Non-negative least squares
-
A type of constrained least squares problem in which the coefficients are restricted to be positive.
- Optimal transport
-
A mathematical framework for finding the most efficient way to transform one probability distribution into another while minimizing a specified cost.
- Segmentation masks
-
The result of a segmentation algorithm, which is used to access and further process the objects identified by the segmentation. In the case of tissue images, the segmentation mask can comprise nuclear and cellular regions, or regions of a certain type (for example, tumour or necrotic regions).
- Spatial statistics
-
A field of applied statistics dealing with spatial data leveraging techniques to study entities using their topological, geometric or geographic properties. Spatial transcriptomics data can be analysed with the latest spatial statistics depending on the layout and features of the technology.
- Topic modelling
-
Unsupervised statistical algorithms originally used in text mining for discovering the latent semantic structures of an extensive text body.
- Tumour nests
-
Small clusters or groups of cancerous cells within a tumour, often surrounded by stromal or immune cells. These nests can have a role in tumour growth, invasion and metastasis.
- Variational autoencoder
-
A generative model that learns probabilistic representations of data by mapping inputs into a latent space (encoding). Variational autoencoders are trained with a pretext task, in which a decoder reconstructs the input data from the latent space representation.
- Vision transformers
-
A deep learning model that applies self-attention mechanisms to image patches, representing an alternative to the widely used convolutional neural networks. Vision transformers achieve high performance in computer vision tasks but require large datasets and substantial computational resources owing to the high number of parameters.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gaspard-Boulinc, L.C., Gortana, L., Walter, T. et al. Cell-type deconvolution methods for spatial transcriptomics. Nat Rev Genet 26, 828–846 (2025). https://doi.org/10.1038/s41576-025-00845-y
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41576-025-00845-y
This article is cited by
-
Advances in Spatial Transcriptomics in Bone
Current Osteoporosis Reports (2026)
-
A guide to transcriptomic deconvolution in cancer
Nature Reviews Cancer (2025)