Abstract
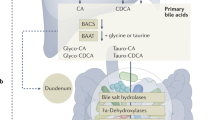
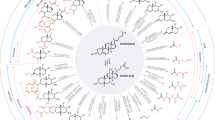
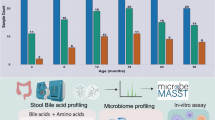
Bile acids are increasingly appearing in the spotlight owing to their novel impacts on various host processes. Similarly, there is growing attention on members of the microbiota that are responsible for bile acid modifications. With recent advances in technology enabling the discovery and continued identification of microbially conjugated bile acids, the chemical complexity of the bile acid landscape in the body is increasing at a rapid pace. In this Review, we summarize our current understanding of how bile acids and the gut microbiota interact to modulate immune responses during homeostasis and disease, with a particular focus on the gut.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Chiang, J. Y. Regulation of bile acid synthesis. Front. Biosci. 3, d176–d193 (1998).
Vlahcevic, Z. R., Pandak, W. M. & Stravitz, R. T. Regulation of bile acid biosynthesis. Gastroenterol. Clin. North. Am. 28, 1–25 (1999).
Jia, W., Xie, G. & Jia, W. Bile acid–microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 15, 111–128 (2018).
Wu, L. et al. The gut microbiome–bile acid axis in hepatocarcinogenesis. Biomed. Pharmacother. 133, 111036 (2021).
Lefebvre, P., Cariou, B., Lien, F., Kuipers, F. & Staels, B. Role of bile acids and bile acid receptors in metabolic regulation. Physiol. Rev. 89, 147–191 (2009).
Ridlon, J. M., Kang, D.-J. & Hylemon, P. B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 47, 241–259 (2006).
Huijghebaert, S. M. & Hofmann, A. F. Influence of the amino acid moiety on deconjugation of bile acid amidates by cholylglycine hydrolase or human fecal cultures. J. Lipid Res. 27, 742–752 (1986).
Dawson, P. A. & Karpen, S. J. Intestinal transport and metabolism of bile acids. J. Lipid Res. 56, 1085–1099 (2015).
Hofmann, A. F. & Hagey, L. R. Key discoveries in bile acid chemistry and biology and their clinical applications: history of the last eight decades. J. Lipid Res. 55, 1553–1595 (2014).
Urdaneta, V. & Casadesús, J. Interactions between bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front. Med. 4, 163 (2017).
Keane, R. M., Gadacz, T. R., Munster, A. M., Birmingham, W. & Winchurch, R. A. Impairment of human lymphocyte function by bile salts. Surgery 95, 439–443 (1984).
Drivas, G., James, O. & Wardle, N. Study of reticuloendothelial phagocytic capacity in patients with cholestasis. Br. Med. J. 1, 1568–1569 (1976).
Podevin, P. et al. Effect of cholestasis and bile acids on interferon-induced 2′,5′-adenylate synthetase and NK cell activities. Gastroenterology 108, 1192–1198 (1995).
Quinn, R. A. et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 579, 123–129 (2020). The identification of a new class of bile acid modifications: microbially conjugated bile acids, produced by the microbiota.
Shalon, D. et al. Profiling the human intestinal environment under physiological conditions. Nature 617, 581–591 (2023).
Zhu, Q.-F. et al. Alternating dual-collision energy scanning mass spectrometry approach: discovery of novel microbial bile-acid conjugates. Anal. Chem. 94, 2655–2664 (2022).
Wang, Y.-Z. et al. A strategy for screening and identification of new amino acid-conjugated bile acids with high coverage by liquid chromatography-mass spectrometry. Anal. Chim. Acta 1239, 340691 (2023).
Pristner, M. et al. Neuroactive metabolites and bile acids are altered in extremely premature infants with brain injury. Cell Rep. Med. 5, 101480 (2024).
Gentry, E. C. et al. Reverse metabolomics for the discovery of chemical structures from humans. Nature 626, 419–426 (2024). Synthesis-based reverse metabolomics led to the identification of new microbial modifications of bile acids.
Wang, H., Chen, J., Hollister, K., Sowers, L. C. & Forman, B. M. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol. Cell 3, 543–553 (1999).
Maruyama, T. et al. Identification of membrane-type receptor for bile acids (M-BAR). Biochem. Biophys. Res. Commun. 298, 714–719 (2002).
Kawamata, Y. et al. A G protein-coupled receptor responsive to bile acids. J. Biol. Chem. 278, 9435–9440 (2003).
Fiorucci, S., Rizzo, G., Donini, A., Distrutti, E. & Santucci, L. Targeting farnesoid X receptor for liver and metabolic disorders. Trends Mol. Med. 13, 298–309 (2007).
Sun, L., Cai, J. & Gonzalez, F. J. The role of farnesoid X receptor in metabolic diseases, and gastrointestinal and liver cancer. Nat. Rev. Gastroenterol. Hepatol. 18, 335–347 (2021).
Sinal, C. J. et al. Targeted disruption of the nuclear receptor FXR/BAR impairs bile acid and lipid homeostasis. Cell 102, 731–744 (2000).
Vassileva, G. et al. Targeted deletion of Gpbar1 protects mice from cholesterol gallstone formation. Biochem. J. 398, 423–430 (2006).
Roberts, L. R. et al. Cathepsin B contributes to bile salt-induced apoptosis of rat hepatocytes. Gastroenterology 113, 1714–1726 (1997).
Rodrigues, C. M., Fan, G., Ma, X., Kren, B. T. & Steer, C. J. A novel role for ursodeoxycholic acid in inhibiting apoptosis by modulating mitochondrial membrane perturbation. J. Clin. Invest. 101, 2790–2799 (1998).
Inagaki, T. et al. Regulation of antibacterial defense in the small intestine by the nuclear bile acid receptor. Proc. Natl Acad. Sci. USA 103, 3920–3925 (2006).
Cipriani, S. et al. The bile acid receptor GPBAR-1 (TGR5) modulates integrity of intestinal barrier and immune response to experimental colitis. PLoS ONE 6, e25637 (2011).
Camilleri, M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut 68, 1516–1526 (2019).
Paray, B. A., Albeshr, M. F., Jan, A. T. & Rather, I. A. Leaky gut and autoimmunity: an intricate balance in individuals health and the diseased state. Int. J. Mol. Sci. 21, 9770 (2020).
Kinashi, Y. & Hase, K. Partners in leaky gut syndrome: intestinal dysbiosis and autoimmunity. Front. Immunol. 12, 673708 (2021).
Seol, W., Choi, H. S. & Moore, D. D. Isolation of proteins that interact specifically with the retinoid X receptor: two novel orphan receptors. Mol. Endocrinol. 9, 72–85 (1995).
Forman, B. M. et al. Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 81, 687–693 (1995).
Bishop-Bailey, D., Walsh, D. T. & Warner, T. D. Expression and activation of the farnesoid X receptor in the vasculature. Proc. Natl Acad. Sci. USA 101, 3668–3673 (2004).
Vavassori, P., Mencarelli, A., Renga, B., Distrutti, E. & Fiorucci, S. The bile acid receptor FXR is a modulator of intestinal innate immunity. J. Immunol. 183, 6251–6261 (2009).
Gadaleta, R. M. et al. Farnesoid X receptor activation inhibits inflammation and preserves the intestinal barrier in inflammatory bowel disease. Gut 60, 463–472 (2011).
Ichikawa, R. et al. Bile acids induce monocyte differentiation toward interleukin-12 hypo-producing dendritic cells via a TGR5-dependent pathway. Immunology 136, 153–162 (2012).
Fiorucci, S., Biagioli, M., Zampella, A. & Distrutti, E. Bile acids activated receptors regulate innate immunity. Front. Immunol. 9, 1853 (2018).
Hofmann, A. F., Hagey, L. R. & Krasowski, M. D. Bile salts of vertebrates: structural variation and possible evolutionary significance. J. Lipid Res. 51, 226–246 (2010).
Russell, D. W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 72, 137–174 (2003).
Chiang, J. Y. L. Bile acids: regulation of synthesis. J. Lipid Res. 50, 1955–1966 (2009).
Takahashi, S. et al. Cyp2c70 is responsible for the species difference in bile acid metabolism between mice and humans. J. Lipid Res. 57, 2130–2137 (2016).
de Boer, J. F. et al. A human-like bile acid pool induced by deletion of hepatic Cyp2c70 modulates effects of FXR activation in mice. J. Lipid Res. 61, 291–305 (2020).
Guo, G. L. & Chiang, J. Y. L. Is CYP2C70 the key to new mouse models to understand bile acids in humans? J. lipid Res. 61, 269–271 (2020).
Honda, A. et al. Regulation of bile acid metabolism in mouse models with hydrophobic bile acid composition. J. Lipid Res. 61, 54–69 (2020).
Falany, C. N., Johnson, M. R., Barnes, S. & Diasio, R. B. Glycine and taurine conjugation of bile acids by a single enzyme. Molecular cloning and expression of human liver bile acid CoA:amino acid N-acyltransferase. J. Biol. Chem. 269, 19375–19379 (1994).
Kirilenko, B. M., Hagey, L. R., Barnes, S., Falany, C. N. & Hiller, M. Evolutionary analysis of bile acid-conjugating enzymes reveals a complex duplication and reciprocal loss history. Genome Biol. Evol. 11, 3256–3268 (2019).
Hofmann, A. F. The continuing importance of bile acids in liver and intestinal disease. Arch. Intern. Med. 159, 2647–2658 (1999).
Hofmann, A. F. & Mysels, K. J. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH, and Ca2+ ions. J. Lipid Res. 33, 617–626 (1992).
Chesney, R. W. et al. The role of taurine in infant nutrition. Adv. Exp. Med. Biol. 442, 463–476 (1998).
Duszka, K. Versatile triad alliance: bile acid, taurine and microbiota. Cells 11, 2337 (2022).
Jones, B. V., Begley, M., Hill, C., Gahan, C. G. M. & Marchesi, J. R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl Acad. Sci. USA 105, 13580–13585 (2008).
Doden, H. L. & Ridlon, J. M. Microbial hydroxysteroid dehydrogenases: from alpha to omega. Microorganisms 9, 469 (2021).
Devendran, S. et al. Clostridium scindens ATCC 35704: integration of nutritional requirements, the complete genome sequence, and global transcriptional responses to bile acids. Appl. Environ. Microbiol. 85, e00052–e00119 (2019).
Wahlström, A., Sayin, S. I., Marschall, H.-U. & Bäckhed, F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab. 24, 41–50 (2016).
Lucas, L. N. et al. Dominant bacterial phyla from the human gut show widespread ability to transform and conjugate bile acids. mSystems https://doi.org/10.1128/msystems.00805-21 (2021).
Yao, L. et al. A biosynthetic pathway for the selective sulfonation of steroidal metabolites by human gut bacteria. Nat. Microbiol. 7, 1404–1418 (2022).
Liu, C. et al. Gut commensal Christensenella minuta modulates host metabolism via acylated secondary bile acids. Nat. Microbiol. 9, 434–450 (2024).
Hofmann, A. F. & Hagey, L. R. Bile acids: chemistry, pathochemistry, biology, pathobiology, and therapeutics. Cell. Mol. Life Sci. 65, 2461–2483 (2008).
Devlin, A. S. & Fischbach, M. A. A biosynthetic pathway for a prominent class of microbiota-derived bile acids. Nat. Chem. Biol. 11, 685–690 (2015).
Paik, D. et al. Human gut bacteria produce ΤΗ17-modulating bile acid metabolites. Nature 603, 907–912 (2022). This study identifies human gut bacteria expressing hydroxysteroid dehydrogenases that convert lithocholic acid to 3-oxolithocholic acid and isolithocholic acid, two bile acids that suppressed TH17 cell differentiation.
Sayin, S. I. et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 17, 225–235 (2013).
Ferrandi, E. E. et al. In search of sustainable chemical processes: cloning, recombinant expression, and functional characterization of the 7α- and 7β-hydroxysteroid dehydrogenases from Clostridium absonum. Appl. Microbiol. Biotechnol. 95, 1221–1233 (2012).
Lee, J.-Y. et al. Contribution of the 7β-hydroxysteroid dehydrogenase from Ruminococcus gnavus N53 to ursodeoxycholic acid formation in the human colon. J. Lipid Res. 54, 3062–3069 (2013).
Song, P. et al. Biological synthesis of ursodeoxycholic acid. Front. Microbiol. 14, 1140662 (2023).
Parks, D. J. et al. Bile acids: natural ligands for an orphan nuclear receptor. Science 284, 1365–1368 (1999).
Jiang, C. et al. Intestine-selective farnesoid X receptor inhibition improves obesity-related metabolic dysfunction. Nat. Commun. 6, 10166 (2015).
Guzior, D. V. et al. Bile salt hydrolase acyltransferase activity expands bile acid diversity. Nature 626, 852–858 (2024). One of the two key studies demonstrating that bile salt hydrolase from members of the gut microbiota has acyltransferase activity and can conjugate amino acids to bile acids.
Rimal, B. et al. Bile salt hydrolase catalyses formation of amine-conjugated bile acids. Nature 626, 859–863 (2024). One of the two key studies demonstrating that bile salt hydrolase from members of the gut microbiota has acyltransferase activity and can conjugate amino acids to bile acids.
Garcia, C. J., Kosek, V., Beltrán, D., Tomás-Barberán, F. A. & Hajslova, J. Production of new microbially conjugated bile acids by human gut microbiota. Biomolecules 12, 687 (2022).
Foley, M. H. et al. Bile salt hydrolases shape the bile acid landscape and restrict Clostridioides difficile growth in the murine gut. Nat. Microbiol. 8, 611–628 (2023).
Folz, J. et al. Human metabolome variation along the upper intestinal tract. Nat. Metab. 5, 777–788 (2023).
Fu, T. et al. Paired microbiome and metabolome analyses associate bile acid changes with colorectal cancer progression. Cell Rep. 42, 112997 (2023).
Curotto de Lafaille, M. A. & Lafaille, J. J. Natural and adaptive foxp3 + regulatory T cells: more of the same or a division of labor? Immunity 30, 626–635 (2009).
van der Veeken, J. et al. Genetic tracing reveals transcription factor Foxp3-dependent and Foxp3-independent functionality of peripherally induced Treg cells. Immunity 55, 1173–1184.e7 (2022).
Ohnmacht, C. et al. Mucosal immunology. The microbiota regulates type 2 immunity through RORγt+ T cells. Science 349, 989–993 (2015).
Sefik, E. et al. Mucosal immunology. Individual intestinal symbionts induce a distinct population of RORγ+ regulatory T cells. Science 349, 993–997 (2015).
Song, X. et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 577, 410–415 (2020). This study shows an essential role for some primary and secondary bile acids and for Bacteroides expressing bile salt hydrolase, in enhancing RORγt+ peripherally induced Treg cells in the colon.
Yang, B.-H. et al. Foxp3+ T cells expressing RORγt represent a stable regulatory T-cell effector lineage with enhanced suppressive capacity during intestinal inflammation. Mucosal Immunol. 9, 444–457 (2016).
Hang, S. et al. Bile acid metabolites control TH17 and Treg cell differentiation. Nature 576, 143–148 (2019). This study demonstrated a key role for bile acids in modulating host immunity by promoting the differentiation of TH17 cells and of Treg cells.
Li, W. et al. A bacterial bile acid metabolite modulates Treg activity through the nuclear hormone receptor NR4A1. Cell Host Microbe 29, 1366–1377.e9 (2021).
Hu, J. et al. Gut microbiota-mediated secondary bile acids regulate dendritic cells to attenuate autoimmune uveitis through TGR5 signaling. Cell Rep. 36, 109726 (2021).
Bettelli, E. et al. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441, 235–238 (2006).
Xiao, R. et al. Synthesis and identification of lithocholic acid 3-sulfate as RORγt ligand to inhibit TH17 cell differentiation. J. Leukoc. Biol. 112, 835–843 (2022).
Campbell, C. et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 581, 475–479 (2020). This study demonstrates that 3β-hydroxydeoxycholic acid modulates antigen-presenting cells to induce colonic RORγt+ peripheral Treg cells.
Keir, M., Yi, T., Lu, T. & Ghilardi, N. The role of IL-22 in intestinal health and disease. J. Exp. Med. 217, e20192195 (2020).
Qi, X. et al. Gut microbiota–bile acid–interleukin-22 axis orchestrates polycystic ovary syndrome. Nat. Med. 25, 1225–1233 (2019). This manuscript describes a role for glycodeoxycholic acid and tauroursodeoxycholic acid in promoting IL-22 production by ileal RORγt+ ILC3s.
Xu, J. et al. An elevated deoxycholic acid level induced by high-fat feeding damages intestinal stem cells by reducing the ileal IL-22. Biochem. Biophys. Res. Commun. 579, 153–160 (2021).
Arifuzzaman, M. et al. Inulin fibre promotes microbiota-derived bile acids and type 2 inflammation. Nature 611, 578–584 (2022). This study links cholic acid and chenodeoxycholic acid to induction of IL-33, increased IL-5 production by ILC2s and enhanced eosinophilia.
Takahashi, S. et al. Role of farnesoid X receptor and bile acids in hepatic tumor development. Hepatol. Commun. 2, 1567–1582 (2018).
Zeng, M. Y., Inohara, N. & Nuñez, G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10, 18–26 (2017).
David, L. A. et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014).
Ocvirk, S. & O’Keefe, S. J. Influence of bile acids on colorectal cancer risk: potential mechanisms mediated by diet–gut microbiota interactions. Curr. Nutr. Rep. 6, 315–322 (2017).
Kosumi, K., Mima, K., Baba, H. & Ogino, S. Dysbiosis of the gut microbiota and colorectal cancer: the key target of molecular pathological epidemiology. J. Lab. Precis. Med. 3, 76 (2018).
Parada Venegas, D. et al. Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10, 277 (2019).
Rogers, A. W. L., Tsolis, R. M. & Bäumler, A. J. Salmonella versus the microbiome. Microbiol. Mol. Biol. Rev. 85, e00027–e00119 (2021).
Litvak, Y. & Bäumler, A. J. Microbiota-nourishing immunity: a guide to understanding our microbial self. Immunity 51, 214–224 (2019).
Byndloss, M. X., Litvak, Y. & Bäumler, A. J. Microbiota-nourishing immunity and its relevance for ulcerative colitis. Inflamm. Bowel Dis. 25, 811–815 (2019).
Miller, B. M. & Bäumler, A. J. The habitat filters of microbiota-nourishing immunity. Annu. Rev. Immunol. 39, 1–18 (2021).
Kelly, C. J. et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671 (2015).
Rivière, A., Selak, M., Lantin, D., Leroy, F. & De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: importance and strategies for their stimulation in the human gut. Front. Microbiol. 7, 979 (2016).
Alex, S. et al. Short-chain fatty acids stimulate angiopoietin-like 4 synthesis in human colon adenocarcinoma cells by activating peroxisome proliferator-activated receptor γ. Mol. Cell. Biol. 33, 1303–1316 (2013).
Byndloss, M. X. et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017).
Litvak, Y., Byndloss, M. X. & Bäumler, A. J. Colonocyte metabolism shapes the gut microbiota. Science 362, eaat9076 (2018).
Fiorucci, S. et al. Cross-talk between farnesoid-X-receptor (FXR) and peroxisome proliferator-activated receptor gamma contributes to the antifibrotic activity of FXR ligands in rodent models of liver cirrhosis. J. Pharmacol. Exp. Ther. 315, 58–68 (2005).
Shinohara, S. & Fujimori, K. Promotion of lipogenesis by PPARγ-activated FXR expression in adipocytes. Biochem. Biophys. Res. Commun. 527, 49–55 (2020).
Abdelkarim, M. et al. The farnesoid X receptor regulates adipocyte differentiation and function by promoting peroxisome proliferator-activated receptor-gamma and interfering with the Wnt/beta-catenin pathways. J. Biol. Chem. 285, 36759–36767 (2010).
Sassone-Corsi, M. & Raffatellu, M. No vacancy: how beneficial microbes cooperate with immunity to provide colonization resistance to pathogens. J. Immunol. 194, 4081–4087 (2015).
Gibold, L. et al. The Vat-AIEC protease promotes crossing of the intestinal mucus layer by Crohn’s disease-associated Escherichia coli. Cell. Microbiol. 18, 617–631 (2016).
Wilson, K. H., Kennedy, M. J. & Fekety, F. R. Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J. Clin. Microbiol. 15, 443–446 (1982).
Thanissery, R., Winston, J. A. & Theriot, C. M. Inhibition of spore germination, growth, and toxin activity of clinically relevant C. difficile strains by gut microbiota derived secondary bile acids. Anaerobe 45, 86–100 (2017).
Sorg, J. A. & Sonenshein, A. L. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J. Bacteriol. 192, 4983–4990 (2010).
Buffie, C. G. et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015).
Tam, J. et al. Intestinal bile acids directly modulate the structure and function of C. difficile TcdB toxin. Proc. Natl Acad. Sci. USA 117, 6792–6800 (2020).
Weingarden, A. R. et al. Ursodeoxycholic acid inhibits Clostridium difficile spore germination and vegetative growth, and prevents the recurrence of ileal pouchitis associated with the infection. J. Clin. Gastroenterol. 50, 624–630 (2016).
Crawford, R. W. et al. Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS Pathog. 8, e1002918 (2012).
Sistrunk, J. R., Nickerson, K. P., Chanin, R. B., Rasko, D. A. & Faherty, C. S. Survival of the fittest: how bacterial pathogens utilize bile to enhance infection. Clin. Microbiol. Rev. 29, 819–836 (2016).
Prouty, A. M., Van Velkinburgh, J. C. & Gunn, J. S. Salmonella enterica serovar Typhimurium resistance to bile: identification and characterization of the tolQRA cluster. J. Bacteriol. 184, 1270–1276 (2002).
Eade, C. R. et al. Bile acids function synergistically to repress invasion gene expression in Salmonella by destabilizing the invasion regulator HilD. Infect. Immun. 84, 2198–2208 (2016).
Yang, X., Stein, K. R. & Hang, H. C. Anti-infective bile acids bind and inactivate a Salmonella virulence regulator. Nat. Chem. Biol. 19, 91–100 (2022).
Basler, M. & Mekalanos, J. J. Type 6 secretion dynamics within and between bacterial cells. Science 337, 815 (2012).
Sana, T. G. et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl Acad. Sci. USA 113, E5044–E5051 (2016).
Gueguen, E. et al. Expression of a yersinia pseudotuberculosis type VI secretion system is responsive to envelope stresses through the OmpR transcriptional activator. PLoS One 8, e66615 (2013).
Bachmann, V. et al. Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl. Trop. Dis. 9, e0004031 (2015).
Lertpiriyapong, K. et al. Campylobacter jejuni type VI secretion system: roles in adaptation to deoxycholic acid, host cell adherence, invasion, and in vivo colonization. PLoS One 7, e42842 (2012).
Matsuoka, K. & Moroi, Y. Micelle formation of sodium deoxycholate and sodium ursodeoxycholate (part 1). Biochim. Biophys. Acta 1580, 189–199 (2002).
Begley, M., Gahan, C. G. M. & Hill, C. The interaction between bacteria and bile. FEMS Microbiol. Rev. 29, 625–651 (2005).
Pedersen, K. J. et al. Eggerthella lenta DSM 2243 alleviates bile acid stress response in Clostridium ramosum and Anaerostipes caccae by transformation of bile acids. Microorganisms 10, 2025 (2022).
Wu, G. D. et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108 (2011).
David, L. A. et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 15, R89 (2014).
Devkota, S. et al. Dietary-fat-induced taurocholic acid promotes pathobiont expansion and colitis in Il10−/− mice. Nature 487, 104–108 (2012).
Carding, S., Verbeke, K., Vipond, D. T., Corfe, B. M. & Owen, L. J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 26, 26191 (2015).
Wotzka, S. Y. et al. Escherichia coli limits Salmonella Typhimurium infections after diet shifts and fat-mediated microbiota perturbation in mice. Nat. Microbiol. 4, 2164–2174 (2019).
Litvak, Y. et al. Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25, 128–139.e5 (2019).
Winter, S. E. et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013).
Caruso, R., Lo, B. C. & Núñez, G. Host–microbiota interactions in inflammatory bowel disease. Nat. Rev. Immunol. 20, 411–426 (2020).
Lloyd-Price, J. et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019).
Hou, R.-G. et al. Bile acid malabsorption is associated with diarrhea in acute phase of colitis. Can. J. Physiol. Pharmacol. 96, 1328–1336 (2018).
Zhou, X. et al. PPARα-UGT axis activation represses intestinal FXR-FGF15 feedback signalling and exacerbates experimental colitis. Nat. Commun. 5, 4573 (2014).
Renga, B. et al. The bile acid sensor FXR is required for immune-regulatory activities of TLR-9 in intestinal inflammation. PLoS ONE 8, e54472 (2013).
Rachmilewitz, D. et al. Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology 122, 1428–1441 (2002).
Rachmilewitz, D. et al. Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 126, 520–528 (2004).
Massafra, V. et al. Splenic dendritic cell involvement in FXR-mediated amelioration of DSS colitis. Biochim. Biophys. Acta 1862, 166–173 (2016).
Fu, T. et al. FXR mediates ILC-intrinsic responses to intestinal inflammation. Proc. Natl Acad. Sci. USA 119, e2213041119 (2022).
Kim, I. et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J. Lipid Res. 48, 2664–2672 (2007).
Goodwin, B. et al. A regulatory cascade of the nuclear receptors FXR, SHP-1, and LRH-1 represses bile acid biosynthesis. Mol. Cell 6, 517–526 (2000).
Kong, B. et al. Mechanism of tissue-specific farnesoid X receptor in suppressing the expression of genes in bile-acid synthesis in mice. Hepatology 56, 1034–1043 (2012).
Ke, J. et al. Fucose ameliorate intestinal inflammation through modulating the crosstalk between bile acids and gut microbiota in a chronic colitis murine model. Inflamm. Bowel Dis. 26, 863–873 (2020).
Vítek, L. Bile acid malabsorption in inflammatory bowel disease. Inflamm. Bowel Dis. 21, 476–483 (2015).
Thomas, J. P., Modos, D., Rushbrook, S. M., Powell, N. & Korcsmaros, T. The emerging role of bile acids in the pathogenesis of inflammatory bowel disease. Front. Immunol. 13, 829525 (2022).
Ridlon, J. M., Daniel, S. L. & Gaskins, H. R. The Hylemon–Björkhem pathway of bile acid 7-dehydroxylation: history, biochemistry, and microbiology. J. Lipid Res. 64, 100392 (2023).
Kim, K. H. et al. Identification and characterization of major bile acid 7α-dehydroxylating bacteria in the human gut. mSystems 7, e0045522 (2022).
Doden, H. L. et al. Completion of the gut microbial epi-bile acid pathway. Gut Microbes 13, 1–20 (2021).
Bai, Y., Zhao, T., Gao, M., Zou, Y. & Lei, X. A novel gene alignment in Dorea sp. AM58-8 produces 7-dehydroxy-3β bile acids from primary bile acids. Biochemistry 61, 2870–2878 (2022).
Dong, Z. & Lee, B. H. Bile salt hydrolases: structure and function, substrate preference, and inhibitor development. Protein Sci. 27, 1742–1754 (2018).
Acknowledgements
M.H.L. was supported by T32 DK007202 and F32 AI169989. M.R. and P.C.D. were funded by National Institutes of Health (NIH) grants AI126277 and DK136117. H.C. was funded by the NIH grant AI167860. M.R. and H.C. were supported by AMED grant JP233fa627003, by the Chiba University-University of California-San Diego (UCSD) Center for Mucosal Immunology, Allergy and Vaccines.
Author information
Authors and Affiliations
Contributions
All authors contributed to the conception and discussion of the article. M.H.L. wrote the first version of the manuscript with contributions from L.R.H. and I.M. M.H.L. and I.M. designed the original concepts for the figures. S.-P.N., P.C.D., H.C. and M.R. provided critical comments. M.H.L., S.-P.N. and M.R. reviewed and edited the manuscript pre-submission and post-submission. All authors approved the submitted version of the article.
Corresponding author
Ethics declarations
Competing interests
P.C.D. consulted for DSM animal health in 2023, is an adviser and holds equity in Cybele, bileOmix and Sirenas and a Scientific co-founder, adviser and holds equity in Ometa, Enveda and Arome with prior approval by UC San Diego. The other authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks F. Schroeder and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Lee, M.H., Nuccio, SP., Mohanty, I. et al. How bile acids and the microbiota interact to shape host immunity. Nat Rev Immunol 24, 798–809 (2024). https://doi.org/10.1038/s41577-024-01057-x
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41577-024-01057-x
This article is cited by
-
Gut dysbiosis in oncology: a risk factor for immunoresistance
Cell Research (2026)
-
Evaluating the efficacy and safety of neoadjuvant immunochemotherapy versus chemotherapy in locally advanced gastric cancer undergoing radical gastrectomy: a retrospective study
World Journal of Surgical Oncology (2025)
-
The interaction between central and peripheral immune systems in methamphetamine use disorder: current status and future directions
Journal of Neuroinflammation (2025)
-
Bile acid metabolism in multiple sclerosis is perturbed and associated with the risk of confirmed disability worsening
BMC Medicine (2025)
-
Endoplasmic reticulum stress responses in anticancer immunity
Nature Reviews Cancer (2025)