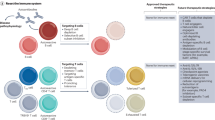
Abstract
Immunoreceptors have crucial roles in sensing environmental signals and initiating immune responses to protect the host. Dysregulation of immunoreceptor signalling can therefore lead to a range of diseases, making immunoreceptor-based therapies a promising frontier in biomedicine. A common feature of various immunoreceptors is the basic-residue-rich sequence (BRS), which is a largely unexplored aspect of immunoreceptor signalling. The BRS is typically located in the cytoplasmic juxtamembrane region of immunoreceptors, where it forms dynamic interactions with neighbouring charged molecules to regulate signalling. Loss or gain of the basic residues in an immunoreceptor BRS has been linked to severe human diseases, such as immunodeficiency and autoimmunity. In this Perspective, we describe the role of BRSs in various immunoreceptors, elucidating their signalling mechanisms and biological functions. Furthermore, we highlight pathogenic mutations in immunoreceptor BRSs and discuss the potential of leveraging BRS signalling in engineered T cell-based therapies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Verdin, P. Top companies and drugs by sales in 2023. Nat. Rev. Drug. Discov. 23, 240 (2024).
June, C. H. & Sadelain, M. Chimeric antigen receptor therapy. N. Engl. J. Med. 379, 64–73 (2018).
Hegde, P. S. & Chen, D. S. Top 10 challenges in cancer immunotherapy. Immunity 52, 17–35 (2020).
Billadeau, D. D. & Leibson, P. J. ITAMs versus ITIMs: striking a balance during cell regulation. J. Clin. Invest. 109, 161–168 (2002).
Yang, W. et al. Dynamic regulation of CD28 conformation and signaling by charged lipids and ions. Nat. Struct. Mol. Biol. 24, 1081–1092 (2017).
Xu, C. et al. Regulation of T cell receptor activation by dynamic membrane binding of the CD3ε cytoplasmic tyrosine-based motif. Cell 135, 702–713 (2008). This paper demonstrates that the electrostatic interactions between the BRS and acidic lipids sequester the adjacent tyrosine motif within the membrane.
Dobbins, J. et al. Binding of the cytoplasmic domain of CD28 to the plasma membrane inhibits lck recruitment and signaling. Sci. Signal. 9, ra75 (2016).
Deford-Watts, L. M. et al. The cytoplasmic tail of the T cell receptor CD3ε subunit contains a phospholipid-binding motif that regulates T cell functions. J. Immunol. 183, 1055–1064 (2009).
Li, L. et al. Ionic CD3–Lck interaction regulates the initiation of T-cell receptor signaling. Proc. Natl Acad. Sci. USA 114, E5891–E5899 (2017).
Wu, W. et al. Multiple signaling roles of CD3ε and its application in CAR-T cell therapy. Cell 182, 855–871 e823 (2020). This paper reports TCR–CD3 phosphorylation patterns and a rational design of CD3ε-based CARs.
Aivazian, D. & Stern, L. J. Phosphorylation of T cell receptor ζ is regulated by a lipid dependent folding transition. Nat. Struct. Biol. 7, 1023–1026 (2000).
Zhang, H., Cordoba, S. P., Dushek, O. & van der Merwe, P. A. Basic residues in the T-cell receptor ζ cytoplasmic domain mediate membrane association and modulate signaling. Proc. Natl Acad. Sci. USA 108, 19323–19328 (2011).
DeFord-Watts, L. M. et al. The CD3 ζ subunit contains a phosphoinositide-binding motif that is required for the stable accumulation of TCR–CD3 complex at the immunological synapse. J. Immunol. 186, 6839–6847 (2011).
Chen, X. et al. Acidic phospholipids govern the enhanced activation of IgG-B cell receptor. Nat. Commun. 6, 8552 (2015).
Wen, M. et al. PD-L1 degradation is regulated by electrostatic membrane association of its cytoplasmic domain. Nat. Commun. 12, 5106 (2021). This paper demonstrates that the electrostatic interactions between the BRS and acidic lipids sequester the lysine ubiquitylation sites within the membrane.
Sarangi, S. K., Lande, K. M. & Kumar, S. KIR signaling is regulated by electrostatic interaction of its cytosolic tail with the plasma membrane despite being neutral polyampholyte. Proc. Natl Acad. Sci. USA 120, e2212987120 (2023).
Cheng, H. et al. Conformational changes in the cytoplasmic region of KIR3DL1 upon interaction with SHP-2. Structure 27, 639–650 e632 (2019).
Sigalov, A. B., Aivazian, D. A., Uversky, V. N. & Stern, L. J. Lipid-binding activity of intrinsically unstructured cytoplasmic domains of multichain immune recognition receptor signaling subunits. Biochemistry 45, 15731–15739 (2006).
Moes, B. et al. INPP5K controls the dynamic structure and signaling of wild-type and mutated, leukemia-associated IL-7 receptors. Blood 141, 1708–1717 (2023).
Dong, R. et al. Molecular dynamics of the recruitment of immunoreceptor signaling module DAP12 homodimer to lipid raft boundary regulated by PIP2. J. Phys. Chem. B 124, 504–510 (2020).
Sun, F. et al. Molecular mechanism for bidirectional regulation of CD44 for lipid raft affiliation by palmitoylations and PIP2. PLoS Comput. Biol. 16, e1007777 (2020).
Araya-Secchi, R. et al. The prolactin receptor scaffolds Janus kinase 2 via co-structure formation with phosphoinositide-4,5-bisphosphate. eLife 12, e84645 (2023).
Haxholm, G. W. et al. Intrinsically disordered cytoplasmic domains of two cytokine receptors mediate conserved interactions with membranes. Biochem. J. 468, 495–506 (2015).
Paddock, C. et al. Residues within a lipid-associated segment of the PECAM-1 cytoplasmic domain are susceptible to inducible, sequential phosphorylation. Blood 117, 6012–6023 (2011).
Wu, W., Shi, X. & Xu, C. Regulation of T cell signalling by membrane lipids. Nat. Rev. Immunol. 16, 690–701 (2016).
Lorent, J. H. et al. Plasma membranes are asymmetric in lipid unsaturation, packing and protein shape. Nat. Chem. Biol. 16, 644–652 (2020).
Doktorova, M., Symons, J. L. & Levental, I. Structural and functional consequences of reversible lipid asymmetry in living membranes. Nat. Chem. Biol. 16, 1321–1330 (2020).
Wen, Y., Vogt, V. M. & Feigenson, G. W. PI(4,5)P2 clustering and its impact on biological functions. Annu. Rev. Biochem. 90, 681–707 (2021).
Posor, Y., Jang, W. & Haucke, V. Phosphoinositides as membrane organizers. Nat. Rev. Mol. Cell Biol. 23, 797–816 (2022).
Schink, K. O., Tan, K. W. & Stenmark, H. Phosphoinositides in control of membrane dynamics. Annu. Rev. Cell Dev. Biol. 32, 143–171 (2016).
Zhukovsky, M. A., Filograna, A., Luini, A., Corda, D. & Valente, C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 593, 2428–2451 (2019).
Golebiewska, U. et al. Membrane-bound basic peptides sequester multivalent (PIP2), but not monovalent (PS), acidic lipids. Biophys. J. 91, 588–599 (2006).
McLaughlin, S. & Murray, D. Plasma membrane phosphoinositide organization by protein electrostatics. Nature 438, 605–611 (2005).
van den Bogaart, G. et al. Membrane protein sequestering by ionic protein–lipid interactions. Nature 479, 552–555 (2011).
Xu, C. et al. A PIP2-derived amplification loop fuels the sustained initiation of B cell activation. Sci. Immunol. 2, eaan0787 (2017).
Wang, J. et al. Growth of B cell receptor microclusters is regulated by PIP2 and PIP3 equilibrium and Dock2 recruitment and activation. Cell Rep. 21, 2541–2557 (2017).
Wan, Z. et al. PI(4,5)P2 determines the threshold of mechanical force-induced B cell activation. J. Cell Biol. 217, 2565–2582 (2018). This paper demonstrates that the electrostatic interactions between the BRSs and PtdIns(4,5)P2 determine the mechanical force threshold required for BCR triggering.
Prakaash, D., Cook, G. P., Acuto, O. & Kalli, A. C. Multi-scale simulations of the T cell receptor reveal its lipid interactions, dynamics and the arrangement of its cytoplasmic region. PLoS Comput. Biol. 17, e1009232 (2021).
Gagnon, E., Schubert, D. A., Gordo, S., Chu, H. H. & Wucherpfennig, K. W. Local changes in lipid environment of TCR microclusters regulate membrane binding by the CD3ε cytoplasmic domain. J. Exp. Med. 209, 2423–2439 (2012).
Chiu, T. Y. et al. INPP5E regulates CD3ζ enrichment at the immune synapse by phosphoinositide distribution control. Commun. Biol. 6, 911 (2023).
Chouaki Benmansour, N. et al. Phosphoinositides regulate the TCR/CD3 complex membrane dynamics and activation. Sci. Rep. 8, 4966 (2018).
Yeung, T. et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science 319, 210–213 (2008).
Zhou, Y. et al. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science 349, 873–876 (2015).
Connolly, A. et al. TMEM16F mediates bystander TCR–CD3 membrane dissociation at the immunological synapse and potentiates T cell activation. Sci. Signal. 14, eabb5146 (2021).
Duchardt, E., Sigalov, A. B., Aivazian, D., Stern, L. J. & Schwalbe, H. Structure induction of the T-cell receptor ζ-chain upon lipid binding investigated by NMR spectroscopy. Chembiochem 8, 820–827 (2007).
Sigalov, A. B. & Hendricks, G. M. Membrane binding mode of intrinsically disordered cytoplasmic domains of T cell receptor signaling subunits depends on lipid composition. Biochem. Biophys. Res. Commun. 389, 388–393 (2009).
Zimmermann, K. et al. The cytosolic domain of T-cell receptor ζ associates with membranes in a dynamic equilibrium and deeply penetrates the bilayer. J. Biol. Chem. 292, 17746–17759 (2017).
Liang, W. et al. Enhancing the antitumor immunity of T cells by engineering the lipid-regulatory site of the TCR/CD3 complex. Cancer Immunol. Res. 11, 93–108 (2023).
Hartl, F. A. et al. Noncanonical binding of Lck to CD3ε promotes TCR signaling and CAR function. Nat. Immunol. 21, 902–913 (2020).
Rudd, C. E. How the discovery of the CD4/CD8–p56(lck) complexes changed immunology and immunotherapy. Front. Cell Dev. Biol. 9, 626095 (2021).
Carmo, A. M., Mason, D. W. & Beyers, A. D. Physical association of the cytoplasmic domain of CD2 with the tyrosine kinases p56lck and p59fyn. Eur. J. Immunol. 23, 2196–2201 (1993).
Raab, M., Yamamoto, M. & Rudd, C. E. The T-cell antigen CD5 acts as a receptor and substrate for the protein-tyrosine kinase p56lck. Mol. Cell Biol. 14, 2862–2870 (1994).
Xu, X. et al. Phase separation of chimeric antigen receptor promotes immunological synapse maturation persistent cytotoxicity. Preprint at SSRN https://doi.org/10.2139/ssrn.4634356 (2023).
Glassman, C. R. et al. Structure of a Janus kinase cytokine receptor complex reveals the basis for dimeric activation. Science 376, 163–169 (2022).
Wilmes, S. et al. Mechanism of homodimeric cytokine receptor activation and dysregulation by oncogenic mutations. Science 367, 643–652 (2020). This paper demonstrates that ligand binding induces a conformational change of the cytoplasmic juxtamembrane region of cytokine receptors.
Brooks, A. J. et al. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science 344, 1249783 (2014).
Bugge, K. et al. A combined computational and structural model of the full-length human prolactin receptor. Nat. Commun. 7, 11578 (2016).
Kassem, N. et al. Order and disorder — an integrative structure of the full-length human growth hormone receptor. Sci. Adv. 7, eabh3805 (2021).
Monks, C. R., Freiberg, B. A., Kupfer, H., Sciaky, N. & Kupfer, A. Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395, 82–86 (1998).
Campi, G., Varma, R. & Dustin, M. L. Actin and agonist mhc-peptide complex-dependent T cell receptor microclusters as scaffolds for signaling. J. Exp. Med. 202, 1031–1036 (2005).
Yokosuka, T. et al. Spatiotemporal regulation of T cell costimulation by TCR–CD28 microclusters and protein kinase C θ translocation. Immunity 29, 589–601 (2008).
Yokosuka, T. et al. Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. J. Exp. Med. 209, 1201–1217 (2012).
Krummel, M. F., Sjaastad, M. D., Wulfing, C. & Davis, M. M. Differential clustering of CD4 and CD3ζ during T cell recognition. Science 289, 1349–1352 (2000).
Liu, W., Wang, H. & Xu, C. Antigen receptor nanoclusters: small units with big functions. Trends Immunol. 37, 680–689 (2016).
Grakoui, A. et al. The immunological synapse: a molecular machine controlling T cell activation. Science 285, 221–227 (1999).
Kuokkanen, E., Sustar, V. & Mattila, P. K. Molecular control of B cell activation and immunological synapse formation. Traffic 16, 311–326 (2015).
Orange, J. S. Formation and function of the lytic NK-cell immunological synapse. Nat. Rev. Immunol. 8, 713–725 (2008).
Feske, S. Calcium signalling in lymphocyte activation and disease. Nat. Rev. Immunol. 7, 690–702 (2007).
Feske, S., Wulff, H. & Skolnik, E. Y. Ion channels in innate and adaptive immunity. Annu. Rev. Immunol. 33, 291–353 (2015).
Liu, C. S. C. et al. Piezo1 mechanosensors optimize human T cell activation. J. Immunol. 200, 1255–1260 (2018).
Solis, A. G. et al. Mechanosensation of cyclical force by PIEZO1 is essential for innate immunity. Nature 573, 69–74 (2019).
Shi, X. et al. Ca2+ regulates T-cell receptor activation by modulating the charge property of lipids. Nature 493, 111–115 (2013). This paper demonstrates that Ca2+ directly neutralizes lipid negative charges, thereby disrupting BRS–lipid interactions.
Han, K., Kim, S. H., Venable, R. M. & Pastor, R. W. Design principles of PI(4,5)P2 clustering under protein-free conditions: specific cation effects and calcium–potassium synergy. Proc. Natl Acad. Sci. USA 119, e2202647119 (2022).
Wang, Y. H. et al. Divalent cation-induced cluster formation by polyphosphoinositides in model membranes. J. Am. Chem. Soc. 134, 3387–3395 (2012).
Zech, T. et al. Accumulation of raft lipids in T-cell plasma membrane domains engaged in TCR signalling. EMBO J. 28, 466–476 (2009).
Ren, Z. et al. Transient hydroxycholesterol treatment restrains TCR signaling to promote long-term immunity. Cell Chem. Biol. 31, 920–931 e926 (2024). This paper demonstrates that 7α-HC and cholesterol regulate BRS–lipid interactions in opposing ways and reports a sterol-based approach for enhancing memory populations in TCR–T cell products.
Chen, H. et al. Self-programmed dynamics of T cell receptor condensation. Proc. Natl Acad. Sci. USA 120, e2217301120 (2023). This paper demonstrates that the electrostatic interactions between the CD3ε BRS and LCK drive liquid–liquid phase separation.
Guy, C. et al. LAG3 associates with TCR–CD3 complexes and suppresses signaling by driving co-receptor–Lck dissociation. Nat. Immunol. 23, 757–767 (2022). This paper reports that the tandem EP motif (glutamic acid–proline-rich tandem repeat) of LAG3 lowers local pH, thereby disrupting LCK interactions with BRS-containing coreceptors.
Latour, S. & Veillette, A. Proximal protein tyrosine kinases in immunoreceptor signaling. Curr. Opin. Immunol. 13, 299–306 (2001).
Veillette, A., Latour, S. & Davidson, D. Negative regulation of immunoreceptor signaling. Annu. Rev. Immunol. 20, 669–707 (2002).
O’Neill, S. K. et al. Monophosphorylation of CD79a and CD79b ITAM motifs initiates a SHIP-1 phosphatase-mediated inhibitory signaling cascade required for B cell anergy. Immunity 35, 746–756 (2011).
Velasco Cardenas, R. M. et al. Harnessing CD3 diversity to optimize CAR T cells. Nat. Immunol. 24, 2135–2149 (2023). This paper exploits different CD3 chains in CAR design and demonstrates the role of the BRS–LCK interaction in CAR signalling.
Kersh, E. N., Shaw, A. S. & Allen, P. M. Fidelity of T cell activation through multistep T cell receptor ζ phosphorylation. Science 281, 572–575 (1998).
Kersh, E. N., Kersh, G. J. & Allen, P. M. Partially phosphorylated T cell receptor ζ molecules can inhibit T cell activation. J. Exp. Med. 190, 1627–1636 (1999).
Zenner, G., Vorherr, T., Mustelin, T. & Burn, P. Differential and multiple binding of signal transducing molecules to the ITAMs of the TCR-ζ chain. J. Cell Biochem. 63, 94–103 (1996).
Ivashkiv, L. B. Cross-regulation of signaling by ITAM-associated receptors. Nat. Immunol. 10, 340–347 (2009).
Gaud, G. et al. CD3ζ ITAMs enable ligand discrimination and antagonism by inhibiting TCR signaling in response to low-affinity peptides. Nat. Immunol. 24, 2121–2134 (2023).
Pfirsch-Maisonnas, S. et al. Inhibitory ITAM signaling traps activating receptors with the phosphatase SHP-1 to form polarized “inhibisome” clusters. Sci. Signal. 4, ra24 (2011).
Morris, R., Kershaw, N. J. & Babon, J. J. The molecular details of cytokine signaling via the JAK/STAT pathway. Protein Sci. 27, 1984–2009 (2018).
Guo, X. et al. Lipid-dependent conformational dynamics underlie the functional versatility of T-cell receptor. Cell Res. 27, 505–525 (2017).
Jin, H., Liu, D., Ni, Y., Wang, H. & Long, D. Quantitative ensemble interpretation of membrane paramagnetic relaxation enhancement (mPRE) for studying membrane-associated intrinsically disordered proteins. J. Am. Chem. Soc. 146, 791–800 (2024).
Wang, H. et al. Probing transient release of membrane-sequestered tyrosine-based signaling motif by solution NMR spectroscopy. J. Phys. Chem. Lett. 8, 3765–3769 (2017).
Hermiston, M. L., Xu, Z. & Weiss, A. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol. 21, 107–137 (2003).
Sasmal, D. K. et al. TCR-pMHC bond conformation controls TCR ligand discrimination. Cell Mol. Immunol. 17, 203–217 (2020).
Alberti, S., Gladfelter, A. & Mittag, T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019).
Rudd, C. E. & Schneider, H. Unifying concepts in CD28, ICOS and CTLA4 co-receptor signalling. Nat. Rev. Immunol. 3, 544–556 (2003).
King, P. D. et al. Analysis of CD28 cytoplasmic tail tyrosine residues as regulators and substrates for the protein tyrosine kinases, EMT and LCK. J. Immunol. 158, 580–590 (1997).
Zhu, C., Chen, W., Lou, J., Rittase, W. & Li, K. Mechanosensing through immunoreceptors. Nat. Immunol. 20, 1269–1278 (2019).
Wan, Z. et al. The activation of IgM- or isotype-switched IgG- and IgE-BCR exhibits distinct mechanical force sensitivity and threshold. eLife 4, e06925 (2015).
Shen, Z. et al. Conformational change within the extracellular domain of B cell receptor in B cell activation upon antigen binding. eLife 8, e42271 (2019).
Engels, N. et al. Recruitment of the cytoplasmic adaptor Grb2 to surface IgG and IgE provides antigen receptor-intrinsic costimulation to class-switched B cells. Nat. Immunol. 10, 1018–1025 (2009).
Kim, S. T. et al. The αβ T cell receptor is an anisotropic mechanosensor. J. Biol. Chem. 284, 31028–31037 (2009).
Liu, B., Chen, W., Evavold, B. D. & Zhu, C. Accumulation of dynamic catch bonds between TCR and agonist peptide-MHC triggers T cell signaling. Cell 157, 357–368 (2014).
Bettini, M. L. et al. Membrane association of the CD3ε signaling domain is required for optimal T cell development and function. J. Immunol. 193, 258–267 (2014).
Duerr, R. H. et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science 314, 1461–1463 (2006). This paper describes a mutation of basic residue in the juxtamembrane region of IL-23R and its association with inflammatory bowel disease.
Momozawa, Y. et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat. Genet. 43, 43–47 (2011).
Di Meglio, P. et al. The IL23R A/Gln381 allele promotes IL-23 unresponsiveness in human memory T-helper 17 cells and impairs Th17 responses in psoriasis patients. J. Invest. Dermatol. 133, 2381–2389 (2013).
Pidasheva, S. et al. Functional studies on the IBD susceptibility gene IL23R implicate reduced receptor function in the protective genetic variant R381Q. PLoS ONE 6, e25038 (2011).
Sarin, R., Wu, X. & Abraham, C. Inflammatory disease protective R381Q IL23 receptor polymorphism results in decreased primary CD4+ and CD8+ human T-cell functional responses. Proc. Natl Acad. Sci. USA 108, 9560–9565 (2011).
Di Meglio, P. et al. The IL23R R381Q gene variant protects against immune-mediated diseases by impairing IL-23-induced Th17 effector response in humans. PLoS ONE 6, e17160 (2011).
Sun, R., Hedl, M. & Abraham, C. IL23 induces IL23R recycling and amplifies innate receptor-induced signalling and cytokines in human macrophages, and the IBD-protective IL23R R381Q variant modulates these outcomes. Gut 69, 264–273 (2020).
Sun, R. & Abraham, C. IL23 promotes antimicrobial pathways in human macrophages, which are reduced with the IBD-protective IL23R R381Q variant. Cell Mol. Gastroenterol. Hepatol. 10, 673–697 (2020).
Kerner, G. et al. Genetic adaptation to pathogens and increased risk of inflammatory disorders in post-neolithic europe. Cell Genom. 3, 100248 (2023). This paper reports the R381Q mutation of IL-23R as a candidate variant for negative selection in European populations.
Shim, J. O. & Seo, J. K. Very early-onset inflammatory bowel disease (IBD) in infancy is a different disease entity from adult-onset IBD; one form of interleukin-10 receptor mutations. J. Hum. Genet. 59, 337–341 (2014).
Begue, B. et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am. J. Gastroenterol. 106, 1544–1555 (2011).
Aschenbrenner, D. et al. Pathogenic interleukin-10 receptor α variants in humans — balancing natural selection and clinical implications. J. Clin. Immunol. 43, 495–511 (2023).
Castigli, E. et al. TACI is mutant in common variable immunodeficiency and IgA deficiency. Nat. Genet. 37, 829–834 (2005).
Salzer, U. et al. Mutations in TNFRSF13B encoding TACI are associated with common variable immunodeficiency in humans. Nat. Genet. 37, 820–828 (2005). Together with the paper by Castigli et al. (2005), this paper reports a loss of basic residue mutation in the juxtamembrane region of TACI and its role in common variable immunodeficiency.
Pan-Hammarstrom, Q. et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat. Genet. 39, 429–430 (2007).
Castigli, E. et al. Reexamining the role of TACI coding variants in common variable immunodeficiency and selective IgA deficiency. Nat. Genet. 39, 430–431 (2007).
de Mattos Barbosa, M. G. et al. TNFRSF13B genotypes control immune-mediated pathology by regulating the functions of innate B cells. JCI Insight 6, e150483 (2021).
Salzer, U. et al. Relevance of biallelic versus monoallelic TNFRSF13B mutations in distinguishing disease-causing from risk-increasing TNFRSF13B variants in antibody deficiency syndromes. Blood 113, 1967–1976 (2009).
Xia, X. Z. et al. TACI is a TRAF-interacting receptor for TALL-1, a tumor necrosis factor family member involved in B cell regulation. J. Exp. Med. 192, 137–143 (2000).
Bacchelli, C. et al. The C76R transmembrane activator and calcium modulator cyclophilin ligand interactor mutation disrupts antibody production and B-cell homeostasis in heterozygous and homozygous mice. J. Allergy Clin. Immunol. 127, 1253–1259 e1213 (2011).
Fried, A. J., Rauter, I., Dillon, S. R., Jabara, H. H. & Geha, R. S. Functional analysis of transmembrane activator and calcium-modulating cyclophilin ligand interactor (TACI) mutations associated with common variable immunodeficiency. J. Allergy Clin. Immunol. 128, 226–228.e221 (2011).
Watanabe-Smith, K. et al. Discovery and functional characterization of a germline, CSF2RB-activating mutation in leukemia. Leukemia 30, 1950–1953 (2016).
Compagno, M. et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature 459, 717–721 (2009).
Alankus, B. et al. Pathological rank signaling in B cells drives autoimmunity and chronic lymphocytic leukemia. J. Exp. Med. 218, e20200517 (2021).
Boggio, E. et al. Mutation of FAS, XIAP, and UNC13D genes in a patient with a complex lymphoproliferative phenotype. Pediatrics 132, e1052–e1058 (2013).
Campagnoli, M. F. et al. The broad spectrum of autoimmune lymphoproliferative disease: molecular bases, clinical features and long-term follow-up in 31 patients. Haematologica 91, 538–541 (2006).
Ruan, W., Lee, C. T. & Desbarats, J. A novel juxtamembrane domain in tumor necrosis factor receptor superfamily molecules activates Rac1 and controls neurite growth. Mol. Biol. Cell 19, 3192–3202 (2008).
Poissonnier, A. et al. CD95-mediated calcium signaling promotes T helper 17 trafficking to inflamed organs in lupus-prone mice. Immunity 45, 209–223 (2016).
Poissonnier, A. et al. Disrupting the CD95-PLCγ1 interaction prevents Th17-driven inflammation. Nat. Chem. Biol. 14, 1079–1089 (2018).
Chen, X. et al. An autoimmune disease variant of IgG1 modulates B cell activation and differentiation. Science 362, 700–705 (2018). This paper reports a gain of basic residue mutation in the cytoplasmic region of human IgG1 and its pathogenic role in autoimmunity.
Yang, B. et al. An Asia-specific variant of human IgG1 represses colorectal tumorigenesis by shaping the tumor microenvironment. J. Clin. Invest. 132, e153454 (2022).
Sun, W. et al. An IGHG1 variant exhibits polarized prevalence and confers enhanced IgG1 antibody responses against life-threatening organisms. Nat. Immunol. 25, 1809–1819 (2024).
Xu, J. et al. Mechanistic insights into the inhibition of a common CTLA-4 gene mutation in the cytoplasmic domain. Molecules 29, 1330 (2024).
Baulu, E., Gardet, C., Chuvin, N. & Depil, S. TCR-engineered T cell therapy in solid tumors: state of the art and perspectives. Sci. Adv. 9, eadf3700 (2023).
Weber, E. W., Maus, M. V. & Mackall, C. L. The emerging landscape of immune cell therapies. Cell 181, 46–62 (2020).
Chan, J. D. et al. Cellular networks controlling T cell persistence in adoptive cell therapy. Nat. Rev. Immunol. 21, 769–784 (2021).
Shah, N. N. & Fry, T. J. Mechanisms of resistance to CAR T cell therapy. Nat. Rev. Clin. Oncol. 16, 372–385 (2019).
Rafiq, S., Hackett, C. S. & Brentjens, R. J. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 17, 147–167 (2020).
Wang, H., Huang, Y. & Xu, C. Charging CAR by electrostatic power. Immunol. Rev. 320, 138–146 (2023).
Chen, J. et al. Tuning charge density of chimeric antigen receptor optimizes tonic signaling and CAR-T cell fitness. Cell Res. 33, 341–354 (2023).
Yang, W. et al. Potentiating the antitumour response of CD8+ T cells by modulating cholesterol metabolism. Nature 531, 651–655 (2016).
Yan, C. et al. Exhaustion-associated cholesterol deficiency dampens the cytotoxic arm of antitumor immunity. Cancer Cell 41, 1276–1293.e1211 (2023).
Waddington, K. E. et al. LXR directly regulates glycosphingolipid synthesis and affects human CD4+ T cell function. Proc. Natl Acad. Sci. USA 118, e2017394118 (2021).
Takahashi, H. et al. Cholesterol 25-hydroxylase is a metabolic switch to constrain T cell-mediated inflammation in the skin. Sci. Immunol. 6, eabb6444 (2021).
Baek, A. E. et al. The cholesterol metabolite 27 hydroxycholesterol facilitates breast cancer metastasis through its actions on immune cells. Nat. Commun. 8, 864 (2017).
Li, M. et al. Enhanced chemo-immunotherapy against melanoma by inhibition of cholesterol esterification in CD8+ T cells. Nanomedicine 14, 2541–2550 (2018).
Pan, J. et al. Potentiation of Kras peptide cancer vaccine by avasimibe, a cholesterol modulator. eBioMedicine 49, 72–81 (2019).
Chen, X., Song, Q., Xia, L. & Xu, X. Synergy of dendritic cell vaccines and avasimibe in treatment of head and neck cancer in mice. Med. Sci. Monit. 23, 4471–4476 (2017).
Zhao, L. et al. Inhibition of cholesterol esterification enzyme enhances the potency of human chimeric antigen receptor T cells against pancreatic carcinoma. Mol. Ther. Oncolyt. 16, 262–271 (2020).
Zhao, L. et al. Cholesterol esterification enzyme inhibition enhances antitumor effects of human chimeric antigen receptors modified T cells. J. Immunother. 41, 45–52 (2018).
Ma, S. et al. Avasimibe can cooperate with a DC-targeting and integration-deficient lentivector to induce stronger HBV specific T cytotoxic response by regulating cholesterol metabolism. Antivir. Res. 216, 105662 (2023).
Hao, M. et al. Combination of metabolic intervention and T cell therapy enhances solid tumor immunotherapy. Sci. Transl. Med. 12, eaaz6667 (2020).
Salter, A. I. et al. Comparative analysis of TCR and CAR signaling informs CAR designs with superior antigen sensitivity and in vivo function. Sci. Signal. 14, eabe2606 (2021).
Roybal, K. T. et al. Engineering T cells with customized therapeutic response programs using synthetic Notch receptors. Cell 167, 419–432.e416 (2016).
Morsut, L. et al. Engineering customized cell sensing and response behaviors using synthetic Notch receptors. Cell 164, 780–791 (2016).
Zhu, I. et al. Modular design of synthetic receptors for programmed gene regulation in cell therapies. Cell 185, 1431–1443.e1416 (2022). This paper reports that the BRS is the best juxtamembrane region for SNIPR activity and tuning BRS charges influences receptor activity.
Jenkins, B. J., Blake, T. J. & Gonda, T. J. Saturation mutagenesis of the β subunit of the human granulocyte-macrophage colony-stimulating factor receptor shows clustering of constitutive mutations, activation of ERK MAP kinase and STAT pathways, and differential β subunit tyrosine phosphorylation. Blood 92, 1989–2002 (1998).
Acknowledgements
C.X. is funded by a Ministry of Science and Technology of the Peoples’s Republic of China (MOST) grant (2023YFA1800200), a Chinese Academy of Sciences (CAS) grant (YSBR-014) and a Science and Technology Commission of Shanghai Municipality (STCSM) grant (23J21901300). X.S. is funded by a MOST grant (2023YFA0915700). We thank H. Wang for discussion and proof-reading.
Author information
Authors and Affiliations
Contributions
C.X. composed the paper. X.S. and X.H. contributed to discussions. C.X. and X.S. wrote the paper and X.H. prepared the figures. X.S. collected the references and performed bioinformatic analyses.
Corresponding authors
Ethics declarations
Competing interests
The authors have patents filed for E-CAR and cholesterol modulation strategies.
Peer review
Peer review information
Nature Reviews Immunology thanks Susana Minguet, who co-reviewed with Nadine Woessner, Wanli Liu, who co-reviewed with Chenguang Xu, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Uniprot: https://www.uniprot.org/
Xu lab: http://xulab.sibcb.ac.cn/
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, X., He, X. & Xu, C. Charge-based immunoreceptor signalling in health and disease. Nat Rev Immunol 25, 298–311 (2025). https://doi.org/10.1038/s41577-024-01105-6
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41577-024-01105-6