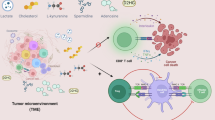
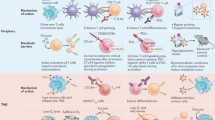
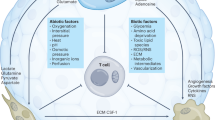
Abstract
Alterations in key metabolic pathways are required for tumour development and the adaptation of tumour cells to intrinsic or extrinsic stresses, as well as for the regulation of immune cell fate and immune responses in the tumour microenvironment. In particular, the dysregulation or alteration of certain metabolites produced by tumour cells has been shown to be important in creating the immunosuppressive tumour microenvironment. Recent studies have broadened our understanding of the interactions between metabolites and antitumour immunity. Here we highlight how, beyond their metabolic role, metabolites can function as signalling molecules to modulate the behaviours of immune cells and tumour cells. We also discuss potential therapeutic strategies targeting specific metabolites and future research directions in metabolite sensing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Mao, Y. & Jiang, P. The crisscross between p53 and metabolism in cancer. Acta Biochim. Biophys. Sin. 55, 914–922 (2023).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Li, L. et al. p53 regulation of ammonia metabolism through urea cycle controls polyamine biosynthesis. Nature 567, 253–256 (2019). The article describes that increased urea cycle flux and polyamine biosynthesis caused by p53 loss are essential for tumor cell proliferation, with implications for p53 mutation-associated tissue pathologies.
Mao, Y., Xia, Z., Xia, W. & Jiang, P. Metabolic reprogramming, sensing, and cancer therapy. Cell Rep. 43, 115064 (2024).
Warburg, O., Wind, F. & Negelein, E. The metabolism of tumors in the body. J. Gen. Physiol. 8, 519–530 (1927).
Liang, J. et al. Selective deficiency of mitochondrial respiratory complex I subunits Ndufs4/6 causes tumor immunogenicity. Nat. Cancer 6, 323–337 (2025).
Cheng, J., Xiao, Y. & Jiang, P. Fumarate integrates metabolism and immunity in diseases. Trends Endocrinol. Metab. https://doi.org/10.1016/j.tem.2025.03.008 (2025).
De Martino, M., Rathmell, J. C., Galluzzi, L. & Vanpouille-Box, C. Cancer cell metabolism and antitumour immunity. Nat. Rev. Immunol. 24, 654–669 (2024).
Cascone, T. et al. Increased tumor glycolysis characterizes immune resistance to adoptive T cell therapy. Cell Metab. 27, 977–987.e974 (2018).
Leone, R. D. et al. Glutamine blockade induces divergent metabolic programs to overcome tumor immune evasion. Science 366, 1013–1021 (2019). This study shows that the use of a glutamine antagonist, JHU083, can simultaneously shut down glycolysis and OXPHOS in mouse cancer cells while increasing OXPHOS in T cells, resulting in them adopting a long-lived, highly activated phenotype.
Reina-Campos, M., Scharping, N. E. & Goldrath, A. W. CD8+ T cell metabolism in infection and cancer. Nat. Rev. Immunol. 21, 718–738 (2021).
Palm, W. & Thompson, C. B. Nutrient acquisition strategies of mammalian cells. Nature 546, 234–242 (2017).
Sousa, C. M. et al. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature 536, 479–483 (2016).
Pucino, V., Bombardieri, M., Pitzalis, C. & Mauro, C. Lactate at the crossroads of metabolism, inflammation, and autoimmunity. Eur. J. Immunol. 47, 14–21 (2017).
Certo, M., Tsai, C. H., Pucino, V., Ho, P. C. & Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 21, 151–161 (2021).
Zhang, W. et al. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell 178, 176–189.e115 (2019).
Comito, G. et al. Lactate modulates CD4+ T-cell polarization and induces an immunosuppressive environment, which sustains prostate carcinoma progression via TLR8/miR21 axis. Oncogene 38, 3681–3695 (2019).
Lygerou, Z., Christophides, G. & Séraphin, B. A novel genetic screen for snRNP assembly factors in yeast identifies a conserved protein, Sad1p, also required for pre-mRNA splicing. Mol. Cell Biol. 19, 2008–2020 (1999).
Ding, R. et al. Lactate modulates RNA splicing to promote CTLA-4 expression in tumor-infiltrating regulatory T cells. Immunity 57, 528–540.e526 (2024).
Lu, S. X. et al. Pharmacologic modulation of RNA splicing enhances anti-tumor immunity. Cell 184, 4032–4047.e4031 (2021).
Kumagai, S. et al. Lactic acid promotes PD-1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell 40, 201–218.e209 (2022).
Feng, Q. et al. Lactate increases stemness of CD8+ T cells to augment anti-tumor immunity. Nat. Commun. 13, 4981 (2022).
Wenes, M. et al. The mitochondrial pyruvate carrier regulates memory T cell differentiation and antitumor function. Cell Metab. 34, 731–746.e739 (2022).
Ranganathan, P. et al. GPR81, a cell-surface receptor for lactate, regulates intestinal homeostasis and protects mice from experimental colitis. J. Immunol. 200, 1781–1789 (2018).
Brown, T. P. et al. The lactate receptor GPR81 promotes breast cancer growth via a paracrine mechanism involving antigen-presenting cells in the tumor microenvironment. Oncogene 39, 3292–3304 (2020).
Roland, C. L. et al. Cell surface lactate receptor GPR81 is crucial for cancer cell survival. Cancer Res. 74, 5301–5310 (2014).
Wagner, W., Kania, K. D., Blauz, A. & Ciszewski, W. M. The lactate receptor (HCAR1/GPR81) contributes to doxorubicin chemoresistance via ABCB1 transporter up-regulation in human cervical cancer HeLa cells. J. Physiol. Pharmacol. 68, 555–564 (2017).
Feng, J. et al. Tumor cell-derived lactate induces TAZ-dependent upregulation of PD-L1 through GPR81 in human lung cancer cells. Oncogene 36, 5829–5839 (2017).
Lundø, K. et al. Lactate receptor GPR81 drives breast cancer growth and invasiveness through regulation of ECM properties and Notch ligand DLL4. BMC Cancer 23, 1136 (2023).
Xing, W., Li, X., Zhou, Y., Li, M. & Zhu, M. Lactate metabolic pathway regulates tumor cell metastasis and its use as a new therapeutic target. Explor. med. 4, 542–560 (2023).
Baryla, M. et al. Oncometabolites-A link between cancer cells and tumor microenvironment. Biology 11, 270 (2022).
Mills, E. & O’Neill, L. A. Succinate: a metabolic signal in inflammation. Trends Cell Biol. 24, 313–320 (2014).
Zhang, Z. et al. Identification of lysine succinylation as a new post-translational modification. Nat. Chem. Biol. 7, 58–63 (2011).
Hirschey, M. D. & Zhao, Y. Metabolic regulation by lysine malonylation, succinylation, and glutarylation. Mol. Cell Proteom. 14, 2308–2315 (2015).
Mangalhara, K. C. et al. Manipulating mitochondrial electron flow enhances tumor immunogenicity. Science 381, 1316–1323 (2023).
Wu, J. Y. et al. Cancer-derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor. Mol. Cell 77, 213–227.e215 (2020).
Trauelsen, M. et al. Extracellular succinate hyperpolarizes M2 macrophages through SUCNR1/GPR91-mediated Gq signaling. Cell Rep. 35, 109246 (2021).
Xiao, Y. et al. Succinate is a natural suppressor of antiviral immune response by targeting MAVS. Front. Immunol. 13, 816378 (2022).
Gudgeon, N. et al. Succinate uptake by T cells suppresses their effector function via inhibition of mitochondrial glucose oxidation. Cell Rep. 40, 111193 (2022).
Rubic, T. et al. Triggering the succinate receptor GPR91 on dendritic cells enhances immunity. Nat. Immunol. 9, 1261–1269 (2008).
Tannahill, G. M. et al. Succinate is an inflammatory signal that induces IL-1beta through HIF-1alpha. Nature 496, 238–242 (2013). This study shows that LPS induces metabolic reprogramming in macrophages, resulting in a marked increase in succinate production, predominantly from glutamine, to stabilize HIF1-α, thereby impairing IFN-β production.
Inamdar, S. et al. Succinate based polymers drive immunometabolism in dendritic cells to generate cancer immunotherapy. J. Control. Rel. 358, 541–554 (2023).
Schmidt, C., Sciacovelli, M. & Frezza, C. Fumarate hydratase in cancer: a multifaceted tumour suppressor. Semin. Cell Dev. Biol. 98, 15–25 (2020).
Yamasaki, T. et al. Exploring a glycolytic inhibitor for the treatment of an FH-deficient type-2 papillary RCC. Nat. Rev. Urol. 8, 165–171 (2011).
Bevan, S. et al. Germline mutations in fumarate hydratase (FH) do not predispose to prostate cancer. Prostate Cancer Prostatic Dis. 6, 12–14 (2003).
Zecchini, V. et al. Fumarate induces vesicular release of mtDNA to drive innate immunity. Nature 615, 499–506 (2023).
Hooftman, A. et al. Macrophage fumarate hydratase restrains mtRNA-mediated interferon production. Nature 615, 490–498 (2023).
Xia, W., Mao, Y., Xia, Z., Cheng, J. & Jiang, P. Metabolic remodelling produces fumarate via the aspartate-argininosuccinate shunt in macrophages as an antiviral defence. Nat. Microbiol. 10, 1115–1129 (2025). Zecchini et al., Hooftman et al. and Xia et al. describe the roles of mitochondrial and cytosolic fumarate in enhancing innate immunity through mechanisms driven by the release of mitochondrial RNA and DNA, and MAVS succination, respectively, to activate interferon production and stimulate an inflammatory response.
Xiao, M. et al. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes. Dev. 26, 1326–1338 (2012).
Duan, Y. et al. ADSL-generated fumarate binds and inhibits STING to promote tumour immune evasion. Nat. Cell Biol. 27, 668–682 (2025).
Zhang, Z., Yang, Y., Chen, Y., Su, J. & Du, W. Malic enzyme 2 maintains metabolic state and anti-tumor immunity of CD8+ T cells. Mol. Cell 84, 3354–3370.e3357 (2024).
Gupta, V. K. et al. Hypoxia-driven oncometabolite L-2HG maintains stemness-differentiation balance and facilitates immune evasion in pancreatic cancer. Cancer Res. 81, 4001–4013 (2021).
Hvinden, I. C., Cadoux-Hudson, T., Schofield, C. J. & McCullagh, J. S. O. Metabolic adaptations in cancers expressing isocitrate dehydrogenase mutations. Cell Rep. Med. 2, 100469 (2021).
Xu, W. et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 19, 17–30 (2011).
Wu, M. J. et al. Mutant IDH1 inhibition induces dsDNA sensing to activate tumor immunity. Science 385, eadl6173 (2024).
Zhao, M. et al. Malic enzyme 2 maintains protein stability of mutant p53 through 2-hydroxyglutarate. Nat. Metab. 4, 225–238 (2022).
Friedrich, M. et al. Tryptophan metabolism drives dynamic immunosuppressive myeloid states in IDH-mutant gliomas. Nat. Cancer 2, 723–740 (2021).
Feng, S. et al. Blockage of L2HGDH-mediated S-2HG catabolism orchestrates macrophage polarization to elicit antitumor immunity. Cell Rep. 43, 114300 (2024).
Bunse, L. et al. Suppression of antitumor T cell immunity by the oncometabolite (R)-2-hydroxyglutarate. Nat. Med. 24, 1192–1203 (2018).
Tyrakis, P. A. et al. S-2-hydroxyglutarate regulates CD8+ T-lymphocyte fate. Nature 540, 236–241 (2016).
Foskolou, I. P., Bunse, L. & Van den Bossche, J. 2-hydroxyglutarate rides the cancer-immunity cycle. Curr. Opin. Biotechnol. 83, 102976 (2023).
Peace, C. G. & O’Neill, L. A. The role of itaconate in host defense and inflammation. J. Clin. Invest. 132, e148548 (2022).
Zhao, H. et al. Myeloid-derived itaconate suppresses cytotoxic CD8+ T cells and promotes tumour growth. Nat. Metab. 4, 1660–1673 (2022).
Lin, H. et al. Itaconate transporter SLC13A3 impairs tumor immunity via endowing ferroptosis resistance. Cancer Cell 42, 2032–2044.e2036 (2024).
Zhao, Y. et al. Neutrophils resist ferroptosis and promote breast cancer metastasis through aconitate decarboxylase 1. Cell Metab. 35, 1688–1703.e1610 (2023).
Kelly, B. & Pearce, E. L. Amino assets: how amino acids support immunity. Cell Metab. 32, 154–175 (2020).
Recouvreux, M. V. et al. Glutamine mimicry suppresses tumor progression through asparagine metabolism in pancreatic ductal adenocarcinoma. Nat. Cancer 5, 100–113 (2024).
Reinfeld, B. I. et al. Cell-programmed nutrient partitioning in the tumour microenvironment. Nature 593, 282–288 (2021). This study shows that in the TME, myeloid cells have the greatest capacity to take up intratumoral glucose, followed by T cells and cancer cells, and cancer cells have the greatest uptake of glutamine; it describes how this distinct nutrient partitioning occurs.
Nakaya, M. et al. Inflammatory T cell responses rely on amino acid transporter ASCT2 facilitation of glutamine uptake and mTORC1 kinase activation. Immunity 40, 692–705 (2014).
Wei, W. et al. Asparagine drives immune evasion in bladder cancer via RIG-I stability and type I IFN signaling. J. Clin. Invest. 135, e186648 (2025).
Wu, J. et al. Asparagine enhances LCK signalling to potentiate CD8+ T-cell activation and anti-tumour responses. Nat. Cell Biol. 23, 75–86 (2021).
Gnanaprakasam, J. N. R. et al. Asparagine restriction enhances CD8+ T cell metabolic fitness and antitumoral functionality through an NRF2-dependent stress response. Nat. Metab. 5, 1423–1439 (2023).
Chang, H. C. et al. Asparagine deprivation enhances T cell antitumour response in patients via ROS-mediated metabolic and signal adaptations. Nat. Metab. 7, 918–927 (2025).
Ala, M. The footprint of kynurenine pathway in every cancer: a new target for chemotherapy. Eur. J. Pharmacol. 896, 173921 (2021).
Cheong, J. E. & Sun, L. Targeting the IDO1/TDO2-KYN-AhR pathway for cancer immunotherapy - challenges and opportunities. Trends Pharmacol. Sci. 39, 307–325 (2018).
Campesato, L. F. et al. Blockade of the AHR restricts a Treg-macrophage suppressive axis induced by L-kynurenine. Nat. Commun. 11, 4011 (2020).
Takenaka, M. C. et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci. 22, 729–740 (2019).
Chen, E. et al. FLI1 promotes IFN-gamma-induced kynurenine production to impair anti-tumor immunity. Nat. Commun. 15, 4590 (2024).
Liu, Y. et al. Tumor-repopulating cells induce PD-1 expression in CD8+ T cells by transferring kynurenine and AhR activation. Cancer Cell 33, 480–494.e487 (2018).
Boadle-Biber, M. C. Regulation of serotonin synthesis. Prog. Biophys. Mol. Biol. 60, 1–15 (1993).
Goralczyk-Binkowska, A., Szmajda-Krygier, D. & Kozlowska, E. The microbiota–gut–brain axis in psychiatric disorders. Int. J. Mol. Sci. 23, 11245 (2022).
Balakrishna, P., George, S., Hatoum, H. & Mukherjee, S. Serotonin pathway in cancer. Int. J. Mol. Sci. 22, 1268 (2021).
Dizeyi, N. et al. Serotonin activates MAP kinase and PI3K/Akt signaling pathways in prostate cancer cell lines. Urol. Oncol-Semin Ori 29, 436–445 (2011).
Jiang, S.-H. et al. Increased serotonin signaling contributes to the warburg effect in pancreatic tumor cells under metabolic stress and promotes growth of pancreatic tumors in mice. Gastroenterology 153, 277–291.e219 (2017).
Sola-Penna, M. et al. Serotonin activates glycolysis and mitochondria biogenesis in human breast cancer cells through activation of the Jak1/STAT3/ERK1/2 and adenylate cyclase/PKA, respectively. Brit J. Cancer 122, 194–208 (2020).
De las Casas-Engel, M. et al. Serotonin skews human macrophage polarization through HTR2B and HTR7. J. Immunol. 190, 2301–2310 (2013).
Cheng, H. H. et al. Control of cyclooxygenase-2 expression and tumorigenesis by endogenous 5-methoxytryptophan. Proc. Natl Acad. Sci. USA 109, 13231–13236 (2012).
Al-Habsi, M. et al. Spermidine activates mitochondrial trifunctional protein and improves antitumor immunity in mice. Science 378, eabj3510 (2022).
Hibino, S. et al. Tumor cell-derived spermidine is an oncometabolite that suppresses TCR clustering for intratumoral CD8+ T cell activation. Proc. Natl Acad. Sci. USA 120, e2305245120 (2023).
Zhu, Y. et al. Cancer cell-derived arginine fuels polyamine biosynthesis in tumor-associated macrophages to promote immune evasion. Cancer Cell 43, 1045–1060.e1047 (2025).
Holbert, C. E., Cullen, M. T., Casero, R. A. Jr. & Stewart, T. M. Polyamines in cancer: integrating organismal metabolism and antitumour immunity. Nat. Rev. Cancer 22, 467–480 (2022).
Murray, P. J., Rathmell, J. & Pearce, E. Snapshot: immunometabolism. Cell Metab. 22, 190–190.e191 (2015).
Salimian Rizi, B. et al. Nitric oxide mediates metabolic coupling of omentum-derived adipose stroma to ovarian and endometrial cancer cells. Cancer Res. 75, 456–471 (2015).
Mao, Y., Shi, D., Li, G. & Jiang, P. Citrulline depletion by ASS1 is required for proinflammatory macrophage activation and immune responses. Mol. Cell 82, 527–541.e527 (2022).
Cane, S., Geiger, R. & Bronte, V. The roles of arginases and arginine in immunity. Nat. Rev. Immunol. 25, 266–284 (2024).
Cui, H. et al. Arg-tRNA synthetase links inflammatory metabolism to RNA splicing and nuclear trafficking via SRRM2. Nat. Cell Biol. 25, 592–603 (2023). This article shows that arginine depletion during inflammation decreases levels of nuclear-localized arginyl-tRNA synthetase (ArgRS) and describes how arginine or ArgRS depletion alters cellular metabolism and peptide–MHC-I presentation.
Geiger, R. et al. L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842.e813 (2016).
Chantranupong, L. et al. The CASTOR proteins are arginine sensors for the mTORC1 pathway. Cell 165, 153–164 (2016).
Soula, M. et al. Glycosphingolipid synthesis mediates immune evasion in KRAS-driven cancer. Nature 633, 451–458 (2024).
Kloosterman, D. J. et al. Macrophage-mediated myelin recycling fuels brain cancer malignancy. Cell 187, 5336–5356.e5330 (2024).
Fan, H. et al. Trans-vaccenic acid reprograms CD8+ T cells and anti-tumour immunity. Nature 623, 1034–1043 (2023).
Nava Lauson, C. B. et al. Linoleic acid potentiates CD8+ T cell metabolic fitness and antitumor immunity. Cell Metab. 35, 633–650.e639 (2023).
Lacher, S. B. et al. PGE2 limits effector expansion of tumour-infiltrating stem-like CD8+ T cells. Nature 629, 417–425 (2024).
Yu, L. et al. Tumor-derived arachidonic acid reprograms neutrophils to promote immune suppression and therapy resistance in triple-negative breast cancer. Immunity 58, 909–925.e907 (2025).
Ping, Y. et al. PD-1 signaling limits expression of phospholipid phosphatase 1 and promotes intratumoral CD8+ T cell ferroptosis. Immunity 57, 2122–2139.e2129 (2024).
Belabed, M. et al. Cholesterol mobilization regulates dendritic cell maturation and the immunogenic response to cancer. Nat. Immunol. 26, 188–199 (2025).
Mukhopadhya, I. & Louis, P. Gut microbiota-derived short-chain fatty acids and their role in human health and disease. Nat. Rev. Microbiol. 23, 635–651 (2025).
Matsushita, M. et al. Gut microbiota-derived short-chain fatty acids promote prostate cancer growth via IGF1 signaling. Cancer Res. 81, 4014–4026 (2021).
Mathewson, N. D. et al. Gut microbiome–derived metabolites modulate intestinal epithelial cell damage and mitigate graft-versus-host disease. Nat. Immunol. 17, 505–513 (2016).
Hu, C. et al. Gut microbiota-derived short-chain fatty acids regulate group 3 innate lymphoid cells in HCC. Hepatology 77, 48–64 (2023).
Luu, M. et al. Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat. Commun. 12, 4077 (2021).
Bachem, A. et al. Microbiota-derived short-chain fatty acids promote the memory potential of antigen-activated CD8+ T cells. Immunity 51, 285–297.e285 (2019).
Zhang, D. et al. Metabolic regulation of gene expression by histone lactylation. Nature 574, 575–580 (2019). This article defines lactate-derived lactylation of histone lysine residues, a phenomenon that can epigenetically modify gene transcription.
Yu, J. et al. Histone lactylation drives oncogenesis by facilitating m6A reader protein YTHDF2 expression in ocular melanoma. Genome Biol. 22, 85 (2021).
Chen, Y. et al. Metabolic regulation of homologous recombination repair by MRE11 lactylation. Cell 187, 294–311.e221 (2024).
Zong, Z. et al. Alanyl-tRNA synthetase, AARS1, is a lactate sensor and lactyltransferase that lactylates p53 and contributes to tumorigenesis. Cell 187, 2375–2392.e2333 (2024).
De Leo, A. et al. Glucose-driven histone lactylation promotes the immunosuppressive activity of monocyte-derived macrophages in glioblastoma. Immunity 57, 1105–1123.e1108 (2024).
Xiong, J. et al. Lactylation-driven METTL3-mediated RNA m(6)A modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 82, 1660–1677.e1610 (2022).
Li, H., Sun, L., Gao, P. & Hu, H. Lactylation in cancer: current understanding and challenges. Cancer Cell 42, 1803–1807 (2024).
Bardella, C. et al. Aberrant succination of proteins in fumarate hydratase-deficient mice and HLRCC patients is a robust biomarker of mutation status. J. Pathol. 225, 4–11 (2011).
Blatnik, M., Frizzell, N., Thorpe, S. R. & Baynes, J. W. Inactivation of glyceraldehyde-3-phosphate dehydrogenase by fumarate in diabetes: formation of S-(2-succinyl)cysteine, a novel chemical modification of protein and possible biomarker of mitochondrial stress. Diabetes 57, 41–49 (2008).
Zheng, L. et al. Fumarate induces redox-dependent senescence by modifying glutathione metabolism. Nat. Commun. 6, 6001 (2015).
Kinch, L., Grishin, N. V. & Brugarolas, J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell 20, 418–420 (2011).
Kornberg, M. D. et al. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360, 449–453 (2018).
Mills, E. L. et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018). This study describes a role for itaconate, production of which can be induced by type I interferons, in alkylating KEAP1 to increase the expression of antioxidant and anti-inflammatory genes upon lipopolysaccharide stimulation in macrophages.
Liao, S. T. et al. 4-Octyl itaconate inhibits aerobic glycolysis by targeting GAPDH to exert anti-inflammatory effects. Nat. Commun. 10, 5091 (2019).
Qin, W. et al. S-glycosylation-based cysteine profiling reveals regulation of glycolysis by itaconate. Nat. Chem. Biol. 15, 983–991 (2019).
Cheng, J. et al. Cancer-cell-derived fumarate suppresses the anti-tumor capacity of CD8+ T cells in the tumor microenvironment. Cell Metab. 35, 961–978.e910 (2023). This study describes a role for tumour cell-derived fumarate accumulation caused by fumarate hydratase deficiency in inhibiting TCR signalling by succinating ZAP70 in tumour-infiltrating CD8+ T cells as a metabolic barrier to CD8+ T cell anti-tumour function.
Sreedhar, A., Wiese, E. K. & Hitosugi, T. Enzymatic and metabolic regulation of lysine succinylation. Genes. Dis. 7, 166–171 (2020).
Li, L. et al. SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat. Commun. 7, 12235 (2016).
Wang, Y. et al. KAT2A coupled with the α-KGDH complex acts as a histone H3 succinyltransferase. Nature 552, 273–277 (2017).
Kurmi, K. et al. Carnitine palmitoyltransferase 1A has a lysine succinyltransferase activity. Cell Rep. 22, 1365–1373 (2018).
Abril, Y. L. N. et al. Pharmacological and genetic perturbation establish SIRT5 as a promising target in breast cancer. Oncogene 40, 1644–1658 (2021).
Park, J. et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol. Cell 50, 919–930 (2013).
Wang, F. et al. SIRT5 desuccinylates and activates pyruvate kinase M2 to block macrophage IL-1β production and to prevent DSS-induced colitis in mice. Cell Rep. 19, 2331–2344 (2017).
Jiang, S. & Yan, W. Succinate in the cancer–immune cycle. Cancer Lett. 390, 45–47 (2017).
Casero, R. A. Jr., Murray Stewart, T. & Pegg, A. E. Polyamine metabolism and cancer: treatments, challenges and opportunities. Nat. Rev. Cancer 18, 681–695 (2018).
Zhou, J. et al. Spermidine-mediated hypusination of translation factor EIF5A improves mitochondrial fatty acid oxidation and prevents non-alcoholic steatohepatitis progression. Nat. Commun. 13, 5202 (2022).
Coni, S. et al. Blockade of EIF5A hypusination limits colorectal cancer growth by inhibiting MYC elongation. Cell Death Dis. 11, 1045 (2020).
Liao, R. et al. AMD1 promotes breast cancer aggressiveness via a spermidine-eIF5A hypusination-TCF4 axis. Breast Cancer Res. 26, 70 (2024).
Puleston, D. J. et al. Polyamine metabolism is a central determinant of helper T cell lineage fidelity. Cell 184, 4186–4202.e4120 (2021).
Puleston, D. J. et al. Polyamines and eIF5A hypusination modulate mitochondrial respiration and macrophage activation. Cell Metab. 30, 352–363.e358 (2019).
Zeng, J. et al. Targeted inhibition of eIF5A(hpu) suppresses tumor growth and polarization of M2-like tumor-associated macrophages in oral cancer. Cell Death Dis. 14, 579 (2023).
Rogowski, K. et al. A family of protein-deglutamylating enzymes associated with neurodegeneration. Cell 143, 564–578 (2010).
Garnham, C. P. et al. Multivalent microtubule recognition by tubulin tyrosine ligase-like family glutamylases. Cell 161, 1112–1123 (2015).
Valenstein, M. L. & Roll-Mecak, A. Graded control of microtubule severing by tubulin glutamylation. Cell 164, 911–921 (2016).
Torrino, S. et al. Mechano-induced cell metabolism promotes microtubule glutamylation to force metastasis. Cell Metab. 33, 1342–1357.e1310 (2021).
Chen, X. et al. The moonlighting function of glutamin synthase 2 promotes immune evasion of pancreatic ductal adenocarcinoma by tubulin tyrosine ligase-like 1-mediated yes1 associated transcriptional regulator glutamylation. Gastroenterology 168, 1137–1152 (2025).
Xia, P. et al. Glutamylation of the DNA sensor cGAS regulates its binding and synthase activity in antiviral immunity. Nat. Immunol. 17, 369–378 (2016).
Liu, B. et al. IL-7Ralpha glutamylation and activation of transcription factor Sall3 promote group 3 ILC development. Nat. Commun. 8, 231 (2017).
Verdin, E. & Ott, M. 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 (2015).
Bhaumik, S. R., Smith, E. & Shilatifard, A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. Mol. Biol. 14, 1008–1016 (2007).
Gu, W. & Roeder, R. G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 90, 595–606 (1997).
Guertin, D. A. & Wellen, K. E. Acetyl-CoA metabolism in cancer. Nat. Rev. Cancer 23, 156–172 (2023).
Chowdhury, S. et al. Intracellular acetyl CoA potentiates the therapeutic efficacy of antitumor CD8+ T cells. Cancer Res. 82, 2640–2655 (2022).
Qiu, J. et al. Acetate promotes T cell effector function during glucose restriction. Cell Rep. 27, 2063–2074.e2065 (2019).
Noe, J. T. et al. Lactate supports a metabolic-epigenetic link in macrophage polarization. Sci. Adv. 7, eabi8602 (2021).
Michishita, E. et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452, 492–496 (2008).
Kingston, R. E. & Narlikar, G. J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Gene Dev. 13, 2339–2352 (1999).
Zou, W. & Green, D. R. Beggars banquet: metabolism in the tumor immune microenvironment and cancer therapy. Cell Metab. 35, 1101–1113 (2023).
Fang, L. et al. Methionine restriction promotes cGAS activation and chromatin untethering through demethylation to enhance antitumor immunity. Cancer Cell 41, 1118–1133.e1112 (2023).
Roy, D. G. et al. Methionine metabolism shapes T helper cell responses through regulation of epigenetic reprogramming. Cell Metab. 31, 250–266.e259 (2020).
Sinclair, L. V. et al. Antigen receptor control of methionine metabolism in T cells. Elife 8, e44210 (2019).
Pandit, M. et al. Methionine consumption by cancer cells drives a progressive upregulation of PD-1 expression in CD4 T cells. Nat. Commun. 14, 2593 (2023).
Bian, Y. et al. Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585, 277–282 (2020).
Rais, R. et al. Discovery of DRP-104, a tumor-targeted metabolic inhibitor prodrug. Sci. Adv. 8, eabq5925 (2022).
Dikshit, A. et al. Potential utility of synthetic D-lactate polymers in skin cancer. JID Innov. 1, 100043 (2021).
Halford, S. et al. A phase I dose-escalation Study of AZD3965, an oral monocarboxylate transporter 1 inhibitor, in patients with advanced cancer. Clin. Cancer Res. 29, 1429–1439 (2023).
Goldberg, F. W. et al. Discovery of clinical candidate AZD0095, a selective inhibitor of monocarboxylate transporter 4 (MCT4) for oncology. J. Med. Chem. 66, 384–397 (2023).
Dhillon, S. Ivosidenib: first global approval. Drugs 78, 1509–1516 (2018).
Stein, E. M. Enasidenib, a targeted inhibitor of mutant IDH2 proteins for treatment of relapsed or refractory acute myeloid leukemia. Future Oncol. 14, 23–40 (2018).
DiNardo, C. D. et al. Glutaminase inhibition in combination with azacytidine in myelodysplastic syndromes: a phase 1b/2 clinical trial and correlative analyses. Nat. Cancer 5, 1515–1533 (2024).
Soth, M. J. et al. Discovery of IPN60090, a clinical stage selective glutaminase-1 (GLS-1) inhibitor with excellent pharmacokinetic and physicochemical properties. J. Med. Chem. 63, 12957–12977 (2020).
Vanauberg, D., Schulz, C. & Lefebvre, T. Involvement of the pro-oncogenic enzyme fatty acid synthase in the hallmarks of cancer: a promising target in anti-cancer therapies. Oncogenesis 12, 16 (2023).
Kelly, W. et al. Phase II investigation of TVB-2640 (Denifanstat) with bevacizumab in patients with first relapse high-grade astrocytoma. Clin. Cancer Res. 29, 2419–2425 (2023).
Cicin, I. et al. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab for advanced urothelial carcinoma: results from the randomized phase III ECHO-303/KEYNOTE-698 study. BMC Cancer 23, 1256 (2024).
Steggerda, S. M. et al. Inhibition of arginase by CB-1158 blocks myeloid cell-mediated immune suppression in the tumor microenvironment. J. Immunother. Cancer 5, 101 (2017).
Ward, P. S. & Thompson, C. B. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21, 297–308 (2012).
Warburg, O. On the origin of cancer cells. Science 123, 309–314 (1956).
Chang, C. H. et al. Metabolic competition in the tumor microenvironment is a driver of cancer progression. Cell 162, 1229–1241 (2015). This article shows that there is competition for glucose between tumour cells and T cells in the TME and describes how glucose consumption by tumours restricts T cells.
Altman, B. J., Stine, Z. E. & Dang, C. V. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 749 (2016).
Koundouros, N. & Poulogiannis, G. Reprogramming of fatty acid metabolism in cancer. Br. J. Cancer 122, 4–22 (2020).
Schiliro, C. & Firestein, B. L. Mechanisms of metabolic reprogramming in cancer cell supporting enhanced growth proliferation. Cell 10, 1056 (2021).
Gubser, P. M. et al. Rapid effector function of memory CD8+ T cells requires an immediate-early glycolytic switch. Nat. Immunol. 14, 1064–1072 (2013).
Pearce, E. L. & Pearce, E. J. Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 (2013).
Van der Windt, G. J. et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012).
Michalek, R. D. et al. Cutting edge: distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011).
Berod, L. et al. De novo fatty acid synthesis controls the fate between regulatory T and T helper 17 cells. Nat. Med. 20, 1327–1333 (2014).
Ikeda, H. et al. Immune evasion through mitochondrial transfer in the tumour microenvironment. Nature 638, 225–236 (2025).
Waters, L. R., Ahsan, F. M., Wolf, D. M., Shirihai, O. & Teitell, M. A. Initial B cell activation induces metabolic reprogramming and mitochondrial remodeling. iScience 5, 99–109 (2018).
Cheng, J. et al. Fumarate suppresses B-cell activation and function through direct inactivation of LYN. Nat. Chem. Biol. 18, 954–962 (2022).
Zhang, B. et al. B cell-derived GABA elicits IL-10+ macrophages to limit anti-tumour immunity. Nature 599, 471–476 (2021).
Rodríguez-Prados, J.-C. et al. Substrate fate in activated macrophages: a comparison between innate, classic, and alternative activation. J. Immunol. 185, 605–614 (2010).
Jha, A. K. et al. Network integration of parallel metabolic and transcriptional data reveals metabolic modules that regulate macrophage polarization. Immunity 42, 419–430 (2015).
Kumar, S. & Dikshit, M. Metabolic insight of neutrophils in health and disease. Front. Immunol. 10, 2099 (2019).
Azevedo, E. P. et al. A metabolic shift toward pentose phosphate pathway is necessary for amyloid fibril-and phorbol 12-myristate 13-acetate-induced neutrophil extracellular trap (NET) formation. J. Biol. Chem. 290, 22174–22183 (2015).
Grant, G. & Ferrer, C. M. The role of the immune tumor microenvironment in shaping metastatic dissemination, dormancy, and outgrowth. Trends Cell Biol. https://doi.org/10.1016/j.tcb.2025.05.006 (2025).
Acknowledgements
The authors thank Z. Xia and Z. Li in the Jiang laboratory for assistance. This study was supported by the National Natural Science Foundation of China (grant nos. 82125030 and 82341022) to P.J., as well as by TsienTang Life Science Development Fund at Tsinghua University. Y.M. was supported by the Postdoctoral Innovation Talent Support Program, the National Natural Science Foundation of China (grant no. 82372824) and the Shuimu Tsinghua Scholar Program.
Author information
Authors and Affiliations
Contributions
P.J. conceived and conceptualized the review scope. Y.M. and W.X. reviewed the literature. Y.M., W.X. and P.J. created the initial versions of the figures. P.J., Y.M. and W.X. wrote the manuscript. P.J. supervised, revised and integrated the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Immunology thanks Christian Frezza, who coreviewed with Desiree Schatton; Ping-Chih Ho; and Cheng-Chin Kuo for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mao, Y., Xia, W. & Jiang, P. Metabolites as signalling molecules in the tumour immune microenvironment. Nat Rev Immunol (2026). https://doi.org/10.1038/s41577-025-01258-y
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41577-025-01258-y