Abstract
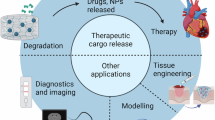
Protein-based biomaterials are growing in popularity for biomedical applications, in part owing to their innate ability to interface with biological systems. These materials, in the form of fibres, nanoparticles and hydrogels, have shown promise as drug delivery vehicles, tissue scaffolds and vaccines. Moreover, the explosion of protein engineering tools and the inception of de novo protein design have transformed our ability to explore new protein structures, enabling the creation of novel materials with diverse properties and furthering their customization for various applications. In this Perspective, we explore the coming of age of protein engineering technologies and their impact on biomaterials. Starting with naturally sourced materials, we highlight common protein building blocks and fabrication methods, as well as recent applications of each. We subsequently explore rationally designed materials and conclude by discussing the potential impacts that de novo design will have on the biomaterials field.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Marin, E., Boschetto, F. & Pezzotti, G. Biomaterials and biocompatibility: an historical overview. J. Biomed. Mater. Res. Part A 108, 1617–1633 (2020).
Bookstaver, M. L., Tsai, S. J., Bromberg, J. S. & Jewell, C. M. Improving vaccine and immunotherapy design using biomaterials. Trends Immunol. 39, 135–150 (2018).
Ratner, B. D. & Bryant, S. J. Biomaterials: where we have been and where we are going. Annu. Rev. Biomed. Eng. 6, 41–75 (2004).
Gomes, S., Leonor, I. B., Mano, J. F., Reis, R. L. & Kaplan, D. L. Natural and genetically engineered proteins for tissue engineering. Prog. Polym. Sci. 37, 1–17 (2012).
Chu, S., Wang, A. L., Bhattacharya, A. & Montclare, J. K. Protein based biomaterials for therapeutic and diagnostic applications. Prog. Biomed. Eng. 4, 012003 (2022).
Catoira, M. C., Fusaro, L., Di Francesco, D., Ramella, M. & Boccafoschi, F. Overview of natural hydrogels for regenerative medicine applications. J. Mater. Sci: Mater. Med. 30, 115 (2019).
Kianfar, E. Protein nanoparticles in drug delivery: animal protein, plant proteins and protein cages, albumin nanoparticles. J. Nanobiotechnol. 19, 159 (2021).
Duan, T., Bian, Q. & Li, H. Light-responsive dynamic protein hydrogels based on LOVTRAP. Langmuir 37, 10214–10222 (2021).
Gregorio, N. E. et al. PhoCoil: a photodegradable and injectable single-component recombinant protein hydrogel for minimally invasive delivery and degradation. Sci. Adv. 11, eadx3472 (2025).
Wang, R., Yang, Z., Luo, J., Hsing, I.-M. & Sun, F. B12-dependent photoresponsive protein hydrogels for controlled stem cell/protein release. Proc. Natl Acad. Sci. USA 114, 5912–5917 (2017).
Jensen, M. M. et al. Temperature-responsive silk-elastin-like protein polymer enhancement of intravesical drug delivery of a therapeutic glycosaminoglycan for treatment of interstitial cystitis/painful bladder syndrome. Biomaterials 217, 119293 (2019). This work demonstrates the utility of silk–elastin-like polypeptide hydrogels as an extended-release delivery vehicle for glycosaminoglycans to the bladder for the treatment of interstitial cystitis, which currently has minimal treatment options that do not provide relief for most patients.
Yang, E. C. et al. Computational design of non-porous pH-responsive antibody nanoparticles. Nat. Struct. Mol. Biol. https://doi.org/10.1038/s41594-024-01288-5 (2024).
Ballister, E. R., Lai, A. H., Zuckermann, R. N., Cheng, Y. & Mougous, J. D. In vitro self-assembly of tailorable nanotubes from a simple protein building block. Proc. Natl Acad. Sci. USA 105, 3733–3738 (2008).
Bian, Q., Kong, N., Arslan, S. & Li, H. Calmodulin-based dynamic protein hydrogels with three distinct mechanical stiffness. Adv. Funct. Mater. 34, 2404934 (2024).
Cao, Y., Wei, X., Lin, Y. & Sun, F. Synthesis of bio-inspired viscoelastic molecular networks by metal-induced protein assembly. Mol. Syst. Des. Eng. 5, 117–124 (2020).
Brodin, J. D., Smith, S. J., Carr, J. R. & Tezcan, F. A. Designed, helical protein nanotubes with variable diameters from a single building block. J. Am. Chem. Soc. 137, 10468–10471 (2015).
Suzuki, Y. et al. Self-assembly of coherently dynamic, auxetic, two-dimensional protein crystals. Nature 533, 369–373 (2016). This study creates 2D self-assembling protein layers by designing specific intermolecular interactions involving metal coordination or disulfide bonds.
Arunachalam, P. S. et al. Durable protection against the SARS-CoV-2 Omicron variant is induced by an adjuvanted subunit vaccine. Sci. Transl. Med. 14, eabq4130 (2022).
Ou, B. S. et al. Broad and durable humoral responses following single hydrogel immunization of SARS-CoV-2 subunit vaccine. Adv. Healthc. Mater. 12, 2301495 (2023).
Ols, S. et al. Multivalent antigen display on nanoparticle immunogens increases B cell clonotype diversity and neutralization breadth to pneumoviruses. Immunity 56, 2425–2441.e14 (2023).
Yuan, Y. et al. Intestinal-targeted nanotubes-in-microgels composite carriers for capsaicin delivery and their effect for alleviation of Salmonella induced enteritis. Biomaterials 287, 121613 (2022). This paper is one of the only published applications of nanotubes to meet a biomedical need, in this case as a delivery system for insoluble drugs such as capsaicin that enable their transport through the mucosal layer for the treatment of enteritis.
Yang, Y. et al. Small molecules combined with collagen hydrogel direct neurogenesis and migration of neural stem cells after spinal cord injury. Biomaterials 269, 120479 (2021). This study demonstrated the efficacy of a collagen hydrogel as a small-molecule delivery vehicle for inducing neurogenesis after spinal cord injury by modulating cell fate at the injury site.
Liu, C., Jiang, T., Yuan, Z. & Lu, Y. Self-assembled casein nanoparticles loading triptolide for the enhancement of oral bioavailability. Nat. Product Commun. https://doi.org/10.1177/1934578X20948352 (2020).
Jiang, B. et al. Injectable, photoresponsive hydrogels for delivering neuroprotective proteins enabled by metal-directed protein assembly. Sci. Adv. 6, eabc4824 (2020).
Maity, B., Samanta, S., Sarkar, S., Alam, S. & Govindaraju, T. Injectable silk fibroin-based hydrogel for sustained insulin delivery in diabetic rats. ACS Appl. Bio Mater. 3, 3544–3552 (2020).
Bennett, J. I. et al. Genetically encoded XTEN-based hydrogels with tunable viscoelasticity and biodegradability for injectable cell therapies. Adv. Sci. https://doi.org/10.1002/advs.202301708 (2024).
Sudhadevi, T., Vijayakumar, H. S., Hariharan, E. V., Sandhyamani, S. & Krishnan, L. K. Optimizing fibrin hydrogel toward effective neural progenitor cell delivery in spinal cord injury. Biomed. Mater. 17, 014102 (2021).
Kortemme, T. De novo protein design — from new structures to programmable functions. Cell 187, 526–544 (2024).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Dauparas, J. et al. Robust deep learning-based protein sequence design using ProteinMPNN. Science 378, 49–56 (2022).
Watson, J. L. et al. De novo design of protein structure and function with RFdiffusion. Nature 620, 1089–1100 (2023). This article demonstrates one of the first examples of use of diffusion-based machine-learning models to generate various protein backbones for many use-cases.
Panahi, R. & Baghban-Salehi, M. in Cellulose-Based Superabsorbent Hydrogels (ed. Mondal, Md. I. H.) 1561–1600 (Springer, 2019).
Liu, B. et al. Protein nanotubes as advanced material platforms and delivery systems. Adv. Mater. 36, 2307627 (2024).
Petrov, A. & Audette, G. F. Peptide and protein-based nanotubes for nanobiotechnology. WIREs Nanomed. Nanobiotechnol. 4, 575–585 (2012).
Audette, G. F., Yaseen, A., Bragagnolo, N. & Bawa, R. Protein nanotubes: from bionanotech towards medical applications. Biomedicines 7, 46 (2019).
Medalsy, I. et al. SP1 protein-based nanostructures and arrays. Nano Lett. 8, 473–477 (2008).
Uddin, I., Frank, S., Warren, M. J. & Pickersgill, R. W. A generic self-assembly process in microcompartments and synthetic protein nanotubes. Small 14, 1704020 (2018).
Shapiro, D. M. et al. Protein nanowires with tunable functionality and programmable self-assembly using sequence-controlled synthesis. Nat. Commun. 13, 829 (2022).
Kumara, M. T., Srividya, N., Muralidharan, S. & Tripp, B. C. Bioengineered flagella protein nanotubes with cysteine loops: self-assembly and manipulation in an optical trap. Nano Lett. 6, 2121–2129 (2006).
Yin, L. et al. Engineered coiled-coil protein for delivery of inverse agonist for osteoarthritis. Biomacromolecules 19, 1614–1624 (2018).
Akhmetova, A. & Heinz, A. Electrospinning proteins for wound healing purposes: opportunities and challenges. Pharmaceutics 13, 4 (2020).
Zhang, C., Zhang, Y., Shao, H. & Hu, X. Hybrid silk fibers dry-spun from regenerated silk fibroin/graphene oxide aqueous solutions. ACS Appl. Mater. Interfaces 8, 3349–3358 (2016).
Rohani Shirvan, A., Nouri, A. & Sutti, A. A perspective on the wet spinning process and its advancements in biomedical sciences. Eur. Polym. J. 181, 111681 (2022).
Kong, B. et al. Tailoring micro/nano-fibers for biomedical applications. Bioact. Mater. 19, 328–347 (2023).
Miranda, C. S. et al. Tunable spun fiber constructs in biomedicine: influence of processing parameters in the fibers’ architecture. Pharmaceutics 14, 164 (2022).
Blackstone, B. N., Gallentine, S. C. & Powell, H. M. Collagen-based electrospun materials for tissue engineering: a systematic review. Bioengineering 8, 39 (2021).
Abrishamkar, A., Nilghaz, A., Saadatmand, M., Naeimirad, M. & deMello, A. J. Microfluidic-assisted fiber production: potentials, limitations, and prospects. Biomicrofluidics 16, 061504 (2022).
Komatsu, T. Protein-based nanotubes for biomedical applications. Nanoscale 4, 1910–1918 (2012).
Landoulsi, J., Roy, C. J., Dupont-Gillain, C. & Demoustier-Champagne, S. Synthesis of collagen nanotubes with highly regular dimensions through membrane-templated layer-by-layer assembly. Biomacromolecules 10, 1021–1024 (2009).
Hou, S., Wang, J. & Martin, C. R. Template-synthesized protein nanotubes. Nano Lett. 5, 231–234 (2005).
Pum, D., Toca-Herrera, J. & Sleytr, U. S-layer protein self-assembly. Int. J. Mol. Sci. 14, 2484–2501 (2013).
Ilk, N., Egelseer, E. M. & Sleytr, U. B. S-layer fusion proteins — construction principles and applications. Curr. Opin. Biotechnol. 22, 824–831 (2011).
Padilla, J. E., Colovos, C. & Yeates, T. O. Nanohedra: using symmetry to design self assembling protein cages, layers, crystals, and filaments. Proc. Natl Acad. Sci. USA 98, 2217–2221 (2001).
Karuppannan, C. et al. Fabrication of progesterone-loaded nanofibers for the drug delivery applications in bovine. Nanoscale Res. Lett. 12, 116 (2017).
Brahatheeswaran, D. et al. Hybrid fluorescent curcumin loaded zein electrospun nanofibrous scaffold for biomedical applications. Biomed. Mater. 7, 045001 (2012).
Nangare, S., Dugam, S., Patil, P., Tade, R. & Jadhav, N. Silk industry waste protein: isolation, purification and fabrication of electrospun silk protein nanofibers as a possible nanocarrier for floating drug delivery. Nanotechnology 32, 035101 (2020).
Xu, L. et al. Mesenchymal stem cell-seeded regenerated silk fibroin complex matrices for liver regeneration in an animal model of acute liver failure. ACS Appl. Mater. Interfaces 9, 14716–14723 (2017).
Xu, H., Cai, S., Sellers, A. & Yang, Y. Intrinsically water-stable electrospun three-dimensional ultrafine fibrous soy protein scaffolds for soft tissue engineering using adipose derived mesenchymal stem cells. RSC Adv. 4, 15451–15457 (2014).
Varshney, N., Sahi, A. K., Poddar, S. & Mahto, S. K. Soy protein isolate supplemented silk fibroin nanofibers for skin tissue regeneration: fabrication and characterization. Int. J. Biol. Macromol. 160, 112–127 (2020).
Zhou, T. et al. Electrospun tilapia collagen nanofibers accelerating wound healing via inducing keratinocytes proliferation and differentiation. Colloids Surf. B Biointerfaces 143, 415–422 (2016).
Bao, C. et al. Enhanced transport of shape and rigidity-tuned α-lactalbumin nanotubes across intestinal mucus and cellular barriers. Nano Lett. 20, 1352–1361 (2020).
Azzaroni, O. & Lau, K. H. A. Layer-by-layer assemblies in nanoporous templates: nano-organized design and applications of soft nanotechnology. Soft Matter 7, 8709–8724 (2011).
Komatsu, T. et al. Virus trap in human serum albumin nanotube. J. Am. Chem. Soc. 133, 3246–3248 (2011).
Yuge, S. et al. Glycoprotein nanotube traps influenza virus. Chem. Lett. 46, 95–97 (2017).
Komatsu, T., Terada, H. & Kobayashi, N. Protein nanotubes with an enzyme interior surface. Chem. Eur. J. 17, 1849–1854 (2011).
Qu, X. & Komatsu, T. Molecular capture in protein nanotubes. ACS Nano 4, 563–573 (2010).
Moll, D. et al. S-layer–streptavidin fusion proteins as template for nanopatterned molecular arrays. Proc. Natl Acad. Sci. USA 99, 14646–14651 (2002).
Riedmann, E. M. et al. Construction of recombinant S-layer proteins (rSbsA) and their expression in bacterial ghosts — a delivery system for the nontypeable Haemophilus influenzae antigen Omp26. FEMS Immunol. Med. Microbiol. 37, 185–192 (2003).
Pleschberger, M. et al. An S-layer heavy chain camel antibody fusion protein for generation of a nanopatterned sensing layer to detect the prostate-specific antigen by surface plasmon resonance technology. Bioconjug. Chem. 15, 664–671 (2004).
Tariq, H., Batool, S., Asif, S., Ali, M. & Abbasi, B. H. Virus-like particles: revolutionary platforms for developing vaccines against emerging infectious diseases. Front. Microbiol. 12, 790121 (2022).
Pretto, C. et al. Cowpea chlorotic mottle virus-like particles as potential platform for antisense oligonucleotide delivery in posterior segment ocular diseases. Macromol. Biosci. 21, 2100095 (2021).
Zhang, J. et al. Advances of structural design and biomedical applications of tobacco mosaic virus coat protein. Adv. NanoBiomed Res. 4, 2300135 (2024).
Hartzell, E. J., Lieser, R. M., Sullivan, M. O. & Chen, W. Modular hepatitis B virus-like particle platform for biosensing and drug delivery. ACS Nano 14, 12642–12651 (2020).
Hashemi, K. et al. Optimizing the synthesis and purification of MS2 virus like particles. Sci. Rep. 11, 19851 (2021).
Chang, J.-Y., Gorzelnik, K. V., Thongchol, J. & Zhang, J. Structural assembly of Qβ virion and its diverse forms of virus-like particles. Viruses 14, 225 (2022).
Cho, K. J. et al. The crystal structure of ferritin from Helicobacter pylori reveals unusual conformational changes for iron uptake. J. Mol. Biol. 390, 83–98 (2009).
Zhang, X., Meining, W., Fischer, M., Bacher, A. & Ladenstein, R. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 Å resolution: determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 306, 1099–1114 (2001).
Van Zon, A., Mossink, M. H., Scheper, R. J., Sonneveld, P. & Wiemer, E. A. C. The vault complex. Cell. Mol. Life Sci. 60, 1828–1837 (2003).
Chmelyuk, N. S., Oda, V. V., Gabashvili, A. N. & Abakumov, M. A. Encapsulins: structure, properties, and biotechnological applications. Biochemistry 88, 35–49 (2023).
Feng, R., Lan, J., Goh, M. C., Du, M. & Chen, Z. Advances in the application of gas vesicles in medical imaging and disease treatment. J. Biol. Eng. 18, 41 (2024).
Yasmin, R., Shah, M., Khan, S. A. & Ali, R. Gelatin nanoparticles: a potential candidate for medical applications. Nanotechnol. Rev. 6, 191–207 (2017).
Pham, D. T. & Tiyaboonchai, W. Fibroin nanoparticles: a promising drug delivery system. Drug Deliv. 27, 431–448 (2020).
Hornok, V. Serum albumin nanoparticles: problems and prospects. Polymers 13, 3759 (2021).
Gandhi, S. & Roy, I. Drug delivery applications of casein nanostructures: a minireview. J. Drug Deliv. Sci. Technol. 66, 102843 (2021).
Lu, L., Duong, V. T., Shalash, A. O., Skwarczynski, M. & Toth, I. Chemical conjugation strategies for the development of protein-based subunit nanovaccines. Vaccines 9, 563 (2021).
Royal, J. M. et al. Development of a SARS-CoV-2 vaccine candidate using plant-based manufacturing and a tobacco mosaic virus-like nano-particle. Vaccines 9, 1347 (2021).
Pomwised, R., Intamaso, U., Teintze, M., Young, M. & Pincus, S. Coupling peptide antigens to virus-like particles or to protein carriers influences the Th1/Th2 polarity of the resulting immune response. Vaccines 4, 15 (2016).
Kanekiyo, M. et al. Self-assembling influenza nanoparticle vaccines elicit broadly neutralizing H1N1 antibodies. Nature 499, 102–106 (2013). This paper demonstrates an early example of fusing an antigen to the self-assembling protein nanoparticle ferritin to develop a potential vaccine.
Jardine, J. et al. Rational HIV immunogen design to target specific germline B cell receptors. Science 340, 711–716 (2013).
Collins, L. T. et al. Encapsulation of AAVs into protein vault nanoparticles as a novel solution to gene therapy’s neutralizing antibody problem. Preprint at bioRxiv https://doi.org/10.1101/2023.11.29.569229 (2024).
Gause, K. T. et al. Immunological principles guiding the rational design of particles for vaccine delivery. ACS Nano 11, 54–68 (2017).
Zhai, L. & Tumban, E. Gardasil-9: a global survey of projected efficacy. Antivir. Res. 130, 101–109 (2016).
Laurens, M. B. RTS,S/AS01 vaccine (MosquirixTM): an overview. Hum. Vaccin. Immunother. 16, 480–489 (2020).
Jardine, J. G. et al. Priming a broadly neutralizing antibody response to HIV-1 using a germline-targeting immunogen. Science 349, 156–161 (2015).
Willis, J. R. et al. Vaccination with mRNA-encoded nanoparticles drives early maturation of HIV bnAb precursors in humans. Science 389, eadr8382 (2025).
Roier, S. et al. mRNA-based VP8* nanoparticle vaccines against rotavirus are highly immunogenic in rodents. npj Vaccines 8, 190 (2023).
Khaleeq, S. et al. Neutralizing efficacy of encapsulin nanoparticles against SARS-CoV2 variants of concern. Viruses 15, 346 (2023).
Hu, X., Cebe, P., Weiss, A. S., Omenetto, F. & Kaplan, D. L. Protein-based composite materials. Mater. Today 15, 208–215 (2012).
Jonker, A. M., Löwik, D. W. P. M. & van Hest, J. C. M. Peptide- and protein-based hydrogels. Chem. Mater. 24, 759–773 (2012).
Silva, N. H. C. S. et al. Protein-based materials: from sources to innovative sustainable materials for biomedical applications. J. Mater. Chem. B 2, 3715–3740 (2014).
Antoine, E. E., Vlachos, P. P. & Rylander, M. N. Review of collagen I hydrogels for bioengineered tissue microenvironments: characterization of mechanics, structure, and transport. Tissue Eng. Part B Rev. 20, 683–696 (2014).
Zhang, H., Xu, D., Zhang, Y., Li, M. & Chai, R. Silk fibroin hydrogels for biomedical applications. Smart Med. 1, e20220011 (2022).
Kong, B. et al. Recombinant human collagen hydrogels with hierarchically ordered microstructures for corneal stroma regeneration. Chem. Eng. J. 428, 131012 (2022).
Arndt, T. et al. Tuneable recombinant spider silk protein hydrogels for drug release and 3D cell culture. Adv. Funct. Mater. 34, 2303622 (2023).
Sindhi, K. et al. The role of biomaterials-based scaffolds in advancing skin tissue construct. J. Tissue Viability 34, 100858 (2025).
Nikolova, M. P. & Chavali, M. S. Recent advances in biomaterials for 3D scaffolds: a review. Bioact. Mater. 4, 271–292 (2019).
Veiga, A., Silva, I. V., Duarte, M. M. & Oliveira, A. L. Current trends on protein driven bioinks for 3D printing. Pharmaceutics 13, 1444 (2021).
Huerta-López, C. & Alegre-Cebollada, J. Protein hydrogels: the Swiss Army Knife for enhanced mechanical and bioactive properties of biomaterials. Nanomaterials 11, 1656 (2021).
He, J., Zhang, N., Zhu, Y., Jin, R. & Wu, F. MSC spheroids-loaded collagen hydrogels simultaneously promote neuronal differentiation and suppress inflammatory reaction through PI3K-Akt signaling pathway. Biomaterials 265, 120448 (2021).
Liu, Y. et al. Construction of adhesive and bioactive silk fibroin hydrogel for treatment of spinal cord injury. Acta Biomater. 158, 178–189 (2023).
Muangsanit, P., Roberton, V., Costa, E. & Phillips, J. B. Engineered aligned endothelial cell structures in tethered collagen hydrogels promote peripheral nerve regeneration. Acta Biomater. 126, 224–237 (2021).
Wei, S.-Y. et al. Engineering large and geometrically controlled vascularized nerve tissue in collagen hydrogels to restore large-sized volumetric muscle loss. Biomaterials 303, 122402 (2023).
Zhang, W. et al. Hydrogel-based dressings designed to facilitate wound healing. Mater. Adv. 5, 1364–1394 (2024).
Li, J., Li, Y., Guo, C. & Wu, X. Development of quercetin loaded silk fibroin/soybean protein isolate hydrogels for burn wound healing. Chem. Eng. J. 481, 148458 (2024).
Jelodari, S. et al. Assessment of the efficacy of an LL-37-encapsulated keratin hydrogel for the treatment of full-thickness wounds. ACS Appl. Bio Mater. 6, 2122–2136 (2023).
Bakadia, B. M. et al. Teicoplanin-decorated reduced graphene oxide incorporated silk protein hybrid hydrogel for accelerating infectious diabetic wound healing and preventing diabetic foot osteomyelitis. Adv. Healthc. Mater. 13, 2304572 (2024).
Gong, W. et al. Construction of a sustained-release hydrogel using gallic acid and lysozyme with antimicrobial properties for wound treatment. Biomater. Sci. 10, 6836–6849 (2022).
Chen, J. et al. Converting lysozyme to hydrogel: a multifunctional wound dressing that is more than antibacterial. Colloids Surf. B Biointerfaces 219, 112854 (2022).
Tian, D.-M. et al. In-situ formed elastin-based hydrogels enhance wound healing via promoting innate immune cells recruitment and angiogenesis. Mater. Today Bio 15, 100300 (2022).
Gagliardi, A. et al. Rutin-loaded zein gel as a green biocompatible formulation for wound healing application. Int. J. Biol. Macromol. 269, 132071 (2024).
Baptista-Silva, S. et al. In situ forming silk sericin-based hydrogel: a novel wound healing biomaterial. ACS Biomater. Sci. Eng. 7, 1573–1586 (2021).
Raza, A. et al. Injectable zein gel with in situ self-assembly as hemostatic material. Biomater. Adv. 145, 213225 (2023).
Tan, J. et al. Biofunctionalized fibrin gel co-embedded with BMSCs and VEGF for accelerating skin injury repair. Mater. Sci. Eng. C 121, 111749 (2021).
Tang, A. et al. Injectable keratin hydrogels as hemostatic and wound dressing materials. Biomater. Sci. 9, 4169–4177 (2021).
Xeroudaki, M. et al. A porous collagen-based hydrogel and implantation method for corneal stromal regeneration and sustained local drug delivery. Sci. Rep. 10, 16936 (2020).
Wang, Y. et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials 128, 44–55 (2017).
McLaughlin, S. et al. Recombinant human collagen hydrogel rapidly reduces methylglyoxal adducts within cardiomyocytes and improves borderzone contractility after myocardial infarction in mice. Adv. Funct. Mater. 32, 2204076 (2022).
Kim, W., Jang, C. H. & Kim, G. H. A myoblast-laden collagen bioink with fully aligned Au nanowires for muscle-tissue regeneration. Nano Lett. 19, 8612–8620 (2019).
Briquez, P. S., Tsai, H.-M., Watkins, E. A. & Hubbell, J. A. Engineered bridge protein with dual affinity for bone morphogenetic protein-2 and collagen enhances bone regeneration for spinal fusion. Sci. Adv. 7, eabh4302 (2021).
Bellas, E. et al. Injectable silk foams for soft tissue regeneration. Adv. Healthc. Mater. 4, 452–459 (2015).
Wang, T. et al. Intraarticularly injectable silk hydrogel microspheres with enhanced mechanical and structural stability to attenuate osteoarthritis. Biomaterials 286, 121611 (2022).
Lee, K. Z. et al. Protein-based hydrogels and their biomedical applications. Molecules 28, 4988 (2023).
Miranda, F. F. et al. A self-assembled protein nanotube with high aspect ratio. Small 5, 2077–2084 (2009).
Hou, C. et al. Construction of protein nanowires through cucurbit[8]uril-based highly specific host–guest interactions: an approach to the assembly of functional proteins. Angew. Chem. Int. Ed. 52, 5590–5593 (2013).
Zhang, X. et al. Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials. Nat. Commun. 12, 4849 (2021).
Brodin, J. D. et al. Metal-directed, chemically tunable assembly of one-, two- and three-dimensional crystalline protein arrays. Nat. Chem. 4, 375–382 (2012).
Sinclair, J. C., Davies, K. M., Vénien-Bryan, C. & Noble, M. E. M. Generation of protein lattices by fusing proteins with matching rotational symmetry. Nat. Nanotechnol. 6, 558–562 (2011).
Gonen, S., DiMaio, F., Gonen, T. & Baker, D. Design of ordered two-dimensional arrays mediated by noncovalent protein–protein interfaces. Science 348, 1365–1368 (2015).
Ben-Sasson, A. J. et al. Design of biologically active binary protein 2D materials. Nature 589, 468–473 (2021).
Tetter, S. et al. Evolution of a virus-like architecture and packaging mechanism in a repurposed bacterial protein. Science 372, 1220–1224 (2021). This work uses principles of directed evolution to create a much larger version of lumazine synthase that can package RNA.
Lai, Y.-T. et al. Structure of a designed protein cage that self-assembles into a highly porous cube. Nat. Chem. 6, 1065–1071 (2014).
Cannon, K. A., Nguyen, V. N., Morgan, C. & Yeates, T. O. Design and characterization of an icosahedral protein cage formed by a double-fusion protein containing three distinct symmetry elements. ACS Synth. Biol. 9, 517–524 (2020).
Lai, Y.-T., Cascio, D. & Yeates, T. O. Structure of a 16-nm cage designed by using protein oligomers. Science 336, 1129 (2012).
Patterson, D. P. et al. Characterization of a highly flexible self-assembling protein system designed to form nanocages. Protein Sci. 23, 190–199 (2014).
Sciore, A. et al. Flexible, symmetry-directed approach to assembling protein cages. Proc. Natl Acad. Sci. USA 113, 8681–8686 (2016).
Badieyan, S. et al. Symmetry-directed self-assembly of a tetrahedral protein cage mediated by de novo-designed coiled coils. ChemBiochem 18, 1888–1892 (2017).
Indelicato, G. et al. Principles governing the self-assembly of coiled-coil protein nanoparticles. Biophys. J. 110, 646–660 (2016).
Fletcher, J. M. et al. Self-assembling cages from coiled-coil peptide modules. Science 340, 595–599 (2013).
Lai, Y.-T., Jiang, L., Chen, W. & Yeates, T. O. On the predictability of the orientation of protein domains joined by a spanning alpha-helical linker. Protein Eng. Des. Sel. 28, 491–499 (2015).
Milligan, J. J., Saha, S., Jenkins, I. C. & Chilkoti, A. Genetically encoded elastin-like polypeptide nanoparticles for drug delivery. Curr. Opin. Biotechnol. 74, 146–153 (2022).
King, N. P. et al. Computational design of self-assembling protein nanomaterials with atomic level accuracy. Science 336, 1171–1174 (2012).
Hsia, Y. et al. Design of a hyperstable 60-subunit protein icosahedron. Nature 535, 136–139 (2016).
Bale, J. B. et al. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science 353, 389–394 (2016).
Wang, J. Y.(J.) et al. Improving the secretion of designed protein assemblies through negative design of cryptic transmembrane domains. Proc. Natl Acad. Sci. USA 120, e2214556120 (2023).
Dowling, Q. M. et al. Hierarchical design of pseudosymmetric protein nanocages. Nature 638, 553–561 (2025).
Lee, S. et al. Four-component protein nanocages designed by programmed symmetry breaking. Nature 638, 546–552 (2025).
Huard, D. J. E., Kane, K. M. & Tezcan, F. A. Re-engineering protein interfaces yields copper-inducible ferritin cage assembly. Nat. Chem. Biol. 9, 169–176 (2013).
Cristie-David, A. S. & Marsh, E. N. G. Metal-dependent assembly of a protein nano-cage. Protein Sci. 28, 1620–1629 (2019).
Golub, E. et al. Constructing protein polyhedra via orthogonal chemical interactions. Nature 578, 172–176 (2020).
Aupič, J. et al. Metal ion-regulated assembly of designed modular protein cages. Sci. Adv. 8, eabm8243 (2022).
Subramanian, R. H. et al. Design of metal-mediated protein assemblies via hydroxamic acid functionalities. Nat. Protoc. 16, 3264–3297 (2021).
Marcandalli, J. et al. Induction of potent neutralizing antibody responses by a designed protein nanoparticle vaccine for respiratory syncytial virus. Cell 176, 1420–1431.e17 (2019).
Brouwer, P. J. M. et al. Enhancing and shaping the immunogenicity of native-like HIV-1 envelope trimers with a two-component protein nanoparticle. Nat. Commun. 10, 4272 (2019).
Ueda, G. et al. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. eLife 9, e57659 (2020).
Song, J. Y. et al. Immunogenicity and safety of SARS-CoV-2 recombinant protein nanoparticle vaccine GBP510 adjuvanted with AS03: interim results of a randomised, active-controlled, observer-blinded, phase 3 trial. eClinicalMedicine 64, 102140 (2023). These results demonstrate the successful use of a computationally designed protein nanoparticle as a viable clinical vaccine candidate in humans.
Hendricks, G. G. et al. Computationally designed mRNA-launched protein nanoparticle immunogens elicit protective antibody and T cell responses in mice. Sci. Transl. Med. 17, eadu2085 (2025).
Walls, A. C. et al. Distinct sensitivities to SARS-CoV-2 variants in vaccinated humans and mice. Cell Rep. 40, 111299 (2022).
Divine, R. et al. Designed proteins assemble antibodies into modular nanocages. Science 372, eabd9994 (2021).
Olshefsky, A. et al. In vivo selection of synthetic nucleocapsids for tissue targeting. Proc. Natl Acad. Sci. USA 120, e2306129120 (2023).
Herpoldt, K. et al. Macromolecular cargo encapsulation via in vitro assembly of two-component protein nanoparticles. Adv. Healthc. Mater. 13, 2303910 (2024).
Edwardson, T. G. W., Mori, T. & Hilvert, D. Rational engineering of a designed protein cage for siRNA delivery. J. Am. Chem. Soc. 140, 10439–10442 (2018).
Garcia Garcia, C., Patkar, S. S., Wang, B., Abouomar, R. & Kiick, K. L. Recombinant protein-based injectable materials for biomedical applications. Adv. Drug Deliv. Rev. 193, 114673 (2023).
Petka, W. A., Harden, J. L., McGrath, K. P., Wirtz, D. & Tirrell, D. A. Reversible hydrogels from self-assembling artificial proteins. Science 281, 389–392 (1998).
Shen, W., Zhang, K., Kornfield, J. A. & Tirrell, D. A. Tuning the erosion rate of artificial protein hydrogels through control of network topology. Nat. Mater. 5, 153–158 (2006).
Olsen, B. D., Kornfield, J. A. & Tirrell, D. A. Yielding behavior in injectable hydrogels from telechelic proteins. Macromolecules 43, 9094–9099 (2010).
Sun, W., Duan, T., Cao, Y. & Li, H. An injectable self-healing protein hydrogel with multiple dissipation modes and tunable dynamic response. Biomacromolecules 20, 4199–4207 (2019).
Meleties, M., Katyal, P., Lin, B., Britton, D. & Montclare, J. K. Self-assembly of stimuli-responsive coiled-coil fibrous hydrogels. Soft Matter 17, 6470–6476 (2021).
Wong Po Foo, C. T. S., Lee, J. S., Mulyasasmita, W., Parisi-Amon, A. & Heilshorn, S. C. Two-component protein-engineered physical hydrogels for cell encapsulation. Proc. Natl Acad. Sci. USA 106, 22067–22072 (2009).
Gao, X., Fang, J., Xue, B., Fu, L. & Li, H. Engineering protein hydrogels using SpyCatcher–SpyTag chemistry. Biomacromolecules 17, 2812–2819 (2016).
Sun, F., Zhang, W.-B., Mahdavi, A., Arnold, F. H. & Tirrell, D. A. Synthesis of bioactive protein hydrogels by genetically encoded SpyTag–SpyCatcher chemistry. Proc. Natl Acad. Sci. USA 111, 11269–11274 (2014).
Lu, Y. et al. Stimuli-responsive protein hydrogels: their design, properties, and biomedical applications. Polymers 2023 15, 4652 (2023).
Lyu, S. et al. Optically controlled reversible protein hydrogels based on photoswitchable fluorescent protein Dronpa. Chem. Commun. 53, 13375–13378 (2017).
Yang, Z. et al. B12-induced reassembly of split photoreceptor protein enables photoresponsive hydrogels with tunable mechanics. Sci. Adv. 8, eabm5482 (2022).
Lao, U. L., Sun, M., Matsumoto, M., Mulchandani, A. & Chen, W. Genetic engineering of self-assembled protein hydrogel based on elastin-like sequences with metal binding functionality. Biomacromolecules 8, 3736–3739 (2007).
Luo, J. & Sun, F. Calcium-responsive hydrogels enabled by inducible protein–protein interactions. Polym. Chem. 11, 4973–4977 (2020).
Lee, J. S., Kang, M. J., Lee, J. H. & Lim, D. W. Injectable hydrogels of stimuli-responsive elastin and calmodulin-based triblock copolypeptides for controlled drug release. Biomacromolecules 23, 2051–2063 (2022).
Wang, Y., Katyal, P. & Montclare, J. K. Protein-engineered functional materials. Adv. Healthc. Mater. 8, 1801374 (2019).
Mout, R. et al. De novo design of modular protein hydrogels with programmable intra- and extracellular viscoelasticity. Proc. Natl Acad. Sci. USA 121, e2309457121 (2024). This paper demonstrates the application of de novo design towards the development of hydrogel materials with variable mechanical properties.
Kong, N., Peng, Q. & Li, H. Rationally designed dynamic protein hydrogels with reversibly tunable mechanical properties. Adv. Funct. Mater. 24, 7310–7317 (2014).
Zhang, Y.-N. et al. A highly elastic and rapidly crosslinkable elastin-like polypeptide-based hydrogel for biomedical applications. Adv. Funct. Mater. 25, 4814–4826 (2015).
Pescador, D. et al. Regeneration of hyaline cartilage promoted by xenogeneic mesenchymal stromal cells embedded within elastin-like recombinamer-based bioactive hydrogels. J. Mater. Sci. Mater. Med. 28, 115 (2017).
Cipriani, F. et al. Cartilage regeneration in preannealed silk elastin-like co-recombinamers injectable hydrogel embedded with mature chondrocytes in an ex vivo culture platform. Biomacromolecules 19, 4333–4347 (2018).
Hatlevik, Ø et al. Translational development of a silk-elastinlike protein polymer embolic for transcatheter arterial embolization. Macromol. Biosci. 22, 2100401 (2022).
Jiang, X. et al. A bi-layer hydrogel cardiac patch made of recombinant functional proteins. Adv. Mater. 34, 2201411 (2022). This paper showcases the power of fusing proteins to design hydrogels with varied properties, in this case to enable the creation of a bilayer material in which the layers have unique functions contributing to their overall utility as a cardiac patch.
Britton, D. et al. Exosome loaded protein hydrogel for enhanced gelation kinetics and wound healing. ACS Appl. Bio Mater. https://doi.org/10.1021/acsabm.4c00569 (2024).
Crick, F. H. C. The Fourier transform of a coiled-coil. Acta Cryst. 6, 685–689 (1953).
Harbury, P. B., Plecs, J. J., Tidor, B., Alber, T. & Kim, P. S. High-resolution protein design with backbone freedom. Science 282, 1462–1467 (1998).
Hill, R. B., Raleigh, D. P., Lombardi, A. & DeGrado, W. F. De novo design of helical bundles as models for understanding protein folding and function. Acc. Chem. Res. 33, 745–754 (2000).
Vázquez Torres, S. et al. De novo designed proteins neutralize lethal snake venom toxins. Nature https://doi.org/10.1038/s41586-024-08393-x (2025).
Huang, B. et al. De novo design of miniprotein antagonists of cytokine storm inducers. Nat. Commun. 15, 7064 (2024).
Shen, H. et al. De novo design of self-assembling helical protein filaments. Science 362, 705–709 (2018).
Shen, H. et al. De novo design of pH-responsive self-assembling helical protein filaments. Nat. Nanotechnol. 19, 1016–1021 (2024). This work exhibits the utility of de novo design to mimic naturally occurring responsive material behaviours that are otherwise difficult to design.
Pyles, H., Zhang, S., De Yoreo, J. J. & Baker, D. Controlling protein assembly on inorganic crystals through designed protein interfaces. Nature 571, 251–256 (2019).
Hsia, Y. et al. Design of multi-scale protein complexes by hierarchical building block fusion. Nat. Commun. 12, 2294 (2021).
Pillai, A. et al. De novo design of allosterically switchable protein assemblies. Nature 632, 911–920 (2024).
Wang, S. et al. Bond-centric modular design of protein assemblies. Nat. Mater. https://doi.org/10.1038/s41563-025-02297-5 (2025).
Rankovic, S. et al. Computational design of bifaceted protein nanomaterials. Nat. Mater. 24, 1635–1643 (2025).
Haas, C. M. et al. From sequence to scaffold: computational design of protein nanoparticle vaccines from AlphaFold2-predicted building blocks. Proc. Natl Acad. Sci. USA 122, e2409566122 (2025).
Gregorio, N. E., Li, Z., Baker, D. & DeForest, C. A. Stimuli-triggered formation of de novo-designed protein biomaterials. Cell Biomater. https://doi.org/10.1016/j.celbio.2025.100239 (2025).
Wicky, B. I. M. et al. Hallucinating symmetric protein assemblies. Science 378, 56–61 (2022).
Chen, Z. et al. Programmable design of orthogonal protein heterodimers. Nature 565, 106–111 (2019).
Cao, L., Zhang, Z., Yuan, D., Yu, M. & Min, J. Tissue engineering applications of recombinant human collagen: a review of recent progress. Front. Bioeng. Biotechnol. 12, 1358246 (2024).
Heinrich, M. A., Mangia, M. & Prakash, J. Impact of endotoxins on bioengineered tissues and models. Trends Biotechnol. 40, 532–534 (2022).
Varadi, M. et al. AlphaFold protein structure database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52, D368–D375 (2024).
Lin, Z. et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 379, 1123–1130 (2023).
Barroca, M. et al. Antibiotic free selection for the high level biosynthesis of a silk-elastin-like protein. Sci. Rep. 6, 39329 (2016).
de Haas, R. J. et al. Rapid and automated design of two-component protein nanomaterials using ProteinMPNN. Proc. Natl Acad. Sci. USA 121, e2314646121 (2024).
Kellmann, S. J. et al. SpyDisplay: a versatile phage display selection system using SpyTag/SpyCatcher technology. mAbs 15, 2177978 (2003).
Baek, M. et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
Dieckhaus, H., Brocidiacono, M., Randolph, N. Z. & Kuhlman, B. Transfer learning to leverage larger datasets for improved prediction of protein stability changes. Proc. Natl Acad. Sci. USA 121, e2314853121 (2024).
Hsu, C. et al. Learning inverse folding from millions of predicted structures. Preprint at bioRxiv https://doi.org/10.1101/2022.04.10.487779 (2022).
Nijkamp, E., Ruffolo, J. A., Weinstein, E. N., Naik, N. & Madani, A. ProGen2: exploring the boundaries of protein language models. Cell Syst. 14, 968–978.e3 (2023).
Ingraham, J. B. et al. Illuminating protein space with a programmable generative model. Nature 623, 1070–1078 (2023).
Acknowledgements
The authors acknowledge support for this work in the form of two Graduate Research Fellowships Program Awards (DGE-2140004 to N.E.G. and C.M.H.) from the National Science Foundation, a Maximizing Investigators’ Research Award (R35GM138036 to C.A.D.) and Other Awards (P01AI167966 and 1U19AI181881 to N.P.K.) from the NIH, and an Award (HR00112420369 to C.A.D.) from the Defense Advanced Research Projects Agency.
Author information
Authors and Affiliations
Contributions
All authors contributed to the writing, editing and review of this manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Materials thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gregorio, N.E., Haas, C.M., King, N.P. et al. Engineering complexity into protein-based biomaterials for biomedical applications. Nat Rev Mater (2025). https://doi.org/10.1038/s41578-025-00861-8
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41578-025-00861-8