Abstract
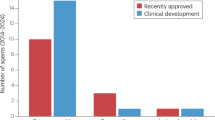
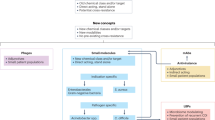
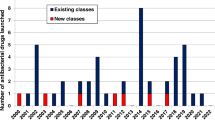
Antibacterial resistance is a great concern and requires global action. A critical question is whether enough new antibacterial drugs are being discovered and developed. A review of the clinical antibacterial drug pipeline was recently published, but comprehensive information about the global preclinical pipeline is unavailable. This Review focuses on discovery and preclinical development projects and has found, as of 1 May 2019, 407 antibacterial projects from 314 institutions. The focus is on Gram-negative pathogens, particularly bacteria on the WHO priority bacteria list. The preclinical pipeline is characterized by high levels of diversity and interesting scientific concepts, with 135 projects on direct-acting small molecules that represent new classes, new targets or new mechanisms of action. There is also a strong trend towards non-traditional approaches, including diverse antivirulence approaches, microbiome-modifying strategies, and engineered phages and probiotics. The high number of pathogen-specific and adjunctive approaches is unprecedented in antibiotic history. Translational hurdles are not adequately addressed yet, especially development pathways to show clinical impact of non-traditional approaches. The innovative potential of the preclinical pipeline compared with the clinical pipeline is encouraging but fragile. Much more work, focus and funding are needed for the novel approaches to result in effective antibacterial therapies to sustainably combat antibacterial resistance.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Lobanovska, M. & Pilla, G. Penicillin's discovery and antibiotic resistance: lessons for the future? Yale J. Biol. Med. 90, 135–145 (2017).
Aminov, R. I. A brief history of the antibiotic era: lessons learned and challenges for the future. Front. Microbiol. 1, 134–134 (2010).
Podolsky, S. H. The antibiotic era: reform, resistance, and the pursuit of a rational therapeutics. (John Hopkins University Press, 2015).
Baker, S. J., Payne, D. J., Rappuoli, R. & De Gregorio, E. Technologies to address antimicrobial resistance. Proc. Natl Acad. Sci. USA 115, 12887–12895 (2018). Extensive review of past antibacterial therapies and opportunities for future strategies to provide new therapies.
Cars, O. Securing access to effective antibiotics for current and future generations. Whose responsibility? Ups J. Med. Sci. 119, 209–214 (2014).
Interagency Coordination Group on Antimicrobial Resistance. No time to wait: securing the future from drug-resistant infections. Report to the Secretary-General of the United Nations. https://www.who.int/antimicrobial-resistance/interagency-coordination-group/IACG_final_report_EN.pdf (2019).
Theuretzbacher, U. et al. Analysis of the clinical antibacterial and antituberculosis pipeline. Lancet Infect. Dis. 19, e40–e50 (2019).
Theuretzbacher, U. & Piddock, L. J. V. Non-traditional antibacterial therapeutic options and challenges. Cell Host Microbe 26, 61–72 (2019).
Wellcome Trust & The Boston Consulting Group. Vaccines to tackle drug resistant infections. An evaluation of R&D opportunities, https://vaccinesforamr.org (2019).
Kortright, K. E., Chan, B. K., Koff, J. L. & Turner, P. E. Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232 (2019).
Motley, M. P. & Fries, B. C. A new take on an old remedy: generating antibodies against multidrug-resistant gram-negative bacteria in a postantibiotic world. mSphere 2, e00397–00317 (2017).
Miró-Canturri, A., Ayerbe-Algaba, R. & Smani, Y. Drug repurposing for the treatment of bacterial and fungal infections. Front Microbiol. 10, 41 (2019).
Rello, J., Parisella, F. R. & Perez, A. Alternatives to antibiotics in an era of difficult-to-treat resistance: new insights. Expert Rev. Clin. Pharmacol. 12, 635–642 (2019).
Monserrat-Martinez, A., Gambin, Y. & Sierecki, E. Thinking outside the bug: molecular targets and strategies to overcome antibiotic resistance. Int. J. Mol. Sci. 20, E1255 (2019). An extensive overview of past and current strategies for antibacterial therapies.
Domalaon, R., Idowu, T., Zhanel, G. G. & Schweizer, F. Antibiotic hybrids: the next generation of agents and adjuvants against gram-negative pathogens? Clin. Microbiol. Rev. 31, e00077–17 (2018).
Kohanski, M. A., Dwyer, D. J. & Collins, J. J. How antibiotics kill bacteria: from targets to networks. Nat. Rev. Microbiol. 8, 423–435 (2010).
Lin, J., Zhou, D., Steitz, T. A., Polikanov, Y. S. & Gagnon, M. G. Ribosome-targeting antibiotics: modes of action, mechanisms of resistance, and implications for drug design. Annu. Rev. Biochem. 87, 451–478 (2018).
Ciumac, D., Gong, H., Hu, X. & Lu, J. R. Membrane targeting cationic antimicrobial peptides. J. Colloid. Interface Sci. 537, 163–185 (2019).
Ma, C., Yang, X. & Lewis, P. J. Bacterial transcription as a target for antibacterial drug development. Microbiol. Mol. Biol. Rev. 80, 139–160 (2016).
Silver, L. L. Appropriate targets for antibacterial drugs. Cold Spring Harb. Perspect. Med. 6, a030239 (2016).
Tacconelli, E. et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327 (2018). The WHO list prioritizes target pathogens for antibacterial drug research and discovery with a global perspective.
Kang, H.-K., Kim, C., Seo, C. H. & Park, Y. The therapeutic applications of antimicrobial peptides (AMPs): a patent review. J. Microbiol. 55, 1–12 (2017).
Molchanova, N., Hansen, P. R. & Franzyk, H. Advances in development of antimicrobial peptidomimetics as potential drugs. Molecules 22, 1430 (2017).
Hoda, M. et al. Antimicrobial peptides: features, action, and their resistance mechanisms in bacteria. Microb. Drug Res. 24, 747–767 (2018).
Torres, M. D. T., Sothiselvam, S., Lu, T. K. & de la Fuente-Nunez, C. Peptide design principles for antimicrobial applications. J. Mol. Biol. 431, 3547–3567 (2019).
Kuppusamy, R., Willcox, M., Black, D. S. & Kumar, N. Short cationic peptidomimetic antimicrobials. Antibiotics 8, 44 (2019).
Wright, G. D. Opportunities for natural products in 21st century antibiotic discovery. Nat. Prod. Rep. 34, 694–701 (2017).
Rossiter, S. E., Fletcher, M. H. & Wuest, W. M. Natural products as platforms to overcome antibiotic resistance. Chem. Rev. 117, 12415–12474 (2017).
Fedorenko, V. et al. Antibacterial discovery and development: from gene to product and back. Biomed. Res. Int. 2015, 591349–591349 (2015).
Erwin, A. L. Antibacterial drug discovery targeting the lipopolysaccharide biosynthetic enzyme LpxC. Cold Spring Harb. Perspect. Med. 6, a025304 (2016).
Fang, L. & Shutao, M. Recent process in the inhibitors of UDP-3-O-(R-3-hydroxyacyl)-nacetylglucosamine deacetylase (LpxC) against gram-negative bacteria. Mini Rev. Med. Chem. 18, 310–323 (2018).
Cohen, F. et al. Optimization of LpxC inhibitors for antibacterial activity and cardiovascular safety. Chem. Med. Chem. 14, 1560–1572 (2019).
US National Library of Medicine. ClinicalTrials.gov, https://clinicaltrials.gov/ct2/show/NCT03832517 (2019).
Wach, A., Dembowsky, K. & Dale, G. E. Pharmacokinetics and safety of intravenous murepavadin infusion in healthy adult subjects administered single and multiple ascending doses. Antimicrob. Agents Chemother. 62, e02355–17 (2018).
Philippon, A., Jacquier, H., Ruppe, E. & Labia, R. Structure-based classification of class a beta-lactamases, an update. Curr. Res. Transl. Med. S2452-3186, 30021–30022 (2019).
Bush, K. & Bradford, P. A. Interplay between β-lactamases and new β-lactamase inhibitors. Nat. Rev. Microbiol. 17, 295–306 (2019).
Shi, C. et al. Approaches for the discovery of metallo-β-lactamase inhibitors: a review. Chem. Biol. Drug Des. 94, 1427–1440 (2019).
Greene, N. P., Kaplan, E., Crow, A. & Koronakis, V. Antibiotic resistance mediated by the MacB ABC transporter family: a structural and functional perspective. Front Microb. 9, 950–950 (2018).
Lamut, A., Peterlin Mašič, L., Kikelj, D. & Tomašič, T. Efflux pump inhibitors of clinically relevant multidrug resistant bacteria. Med. Res. Rev. https://doi.org/10.1002/med.21591 (2019).
Peyclit, L., Baron, S. A. & Rolain, J.-M. Drug repurposing to fight colistin and carbapenem-resistant bacteria. Front Cell Infect. Microbiol. 9, 193–193 (2019).
Rohde, C., Wittmann, J. & Kutter, E. Bacteriophages: a therapy concept against multi-drug-resistant bacteria. Surg Infect. 19, 737–744 (2018).
McCallin, S., Sacher, J. C., Zheng, J. & Chan, B. K. Current state of compassionate phage therapy. Viruses 11, 343 (2019).
Morozova, V. V., Vlassov, V. V. & Tikunova, N. V. Applications of bacteriophages in the treatment of localized infections in humans. Front. Microbiol. 9, 1696–1696 (2018).
Chang, R. Y. K. et al. Phage therapy for respiratory infections. Adv. Drug Deliv. Rev. 133, 76–86 (2018).
Dąbrowska, K. Phage therapy: what factors shape phage pharmacokinetics and bioavailability? Systematic and critical review. Med. Res. Rev. https://doi.org/10.1002/med.21572 (2019). Critical challenges in treating bacterial infections at different body sites are reviewed.
Hatoum-Aslan, A. Phage genetic engineering using CRISPR–Cas systems. Viruses 10, 335 (2018).
Dams, D., Brøndsted, L., Drulis-Kawa, Z. & Briers, Y. Engineering of receptor-binding proteins in bacteriophages and phage tail-like bacteriocins. Biochem. Soc. Trans. 47, 449–460 (2019).
Patsali, P., Kleanthous, M. & Lederer, C. W. Disruptive technology: CRISPR/Cas-based tools and approaches. Mol. Diagn. Ther. 23, 187–200 (2019).
Vila, J. Microbiota transplantation and/or CRISPR/Cas in the battle against antimicrobial resistance. Clin. Microbiol. Infect. 24, 684–686 (2018).
Segall, A. M., Roach, D. R. & Strathdee, S. A. Stronger together? Perspectives on phage-antibiotic synergy in clinical applications of phage therapy. Curr. Opin. Microbiol. 51, 46–50 (2019).
Rex, J. H., Fernandez Lynch, H., Cohen, G., Darrow, J. J. & Outterson, K. Designing development programs for non-traditional antibacterial agents. Nat. Commun. 10, 3416 (2019).
Fischetti, V. A. Development of phage lysins as novel therapeutics: a historical perspective. Viruses 10, 310 (2018).
Abdelkader, K., Gerstmans, H., Saafan, A., Dishisha, T. & Briers, Y. The preclinical and clinical progress of bacteriophages and their lytic enzymes: the parts are easier than the whole. Viruses 11, 96 (2019).
Oliveira, H., São-José, C. & Azeredo, J. Phage-derived peptidoglycan degrading enzymes: challenges and future prospects for in vivo therapy. Viruses 10, 292 (2018).
Briers, Y. & Lavigne, R. Breaking barriers: expansion of the use of endolysins as novel antibacterials against gram-negative bacteria. Future Microbiol. 10, 377–390 (2015).
Falony, G. et al. The human microbiome in health and disease: hype or hope. Acta Clin. Belg. 74, 53–64 (2019). Recent gut microbiome research may help to accelerate translation of microbiome findings from bench to bedside.
Cammarota, G. et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 66, 569–580 (2017).
FDA. FDA In Brief: FDA warns about potential risk of serious infections caused by multi-drug resistant organisms related to the investigational use of fecal microbiota for transplantation. https://www.fda.gov/news-events/fda-brief/fda-brief-fda-warns-about-potential-risk-serious-infections-caused-multi-drug-resistant-organisms (2019).
Kellermayer, R. Fecal microbiota transplantation: great potential with many challenges. Transl. Gastroenterol. Hepatol. 4, 40–40 (2019).
Iqbal, U., Anwar, H. & Karim, M. A. Safety and efficacy of encapsulated fecal microbiota transplantation for recurrent clostridium difficile infection: a systematic review. Eur. J. Gastroenterol. Hepatol. 30, 730–734 (2018).
Mathipa, M. G. & Thantsha, M. S. Probiotic engineering: towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens. Gut Pathog. 9, 28–28 (2017).
Ghosh, C., Sarkar, P., Issa, R. & Haldar, J. Alternatives to conventional antibiotics in the era of antimicrobial resistance. Trends Microbiol. 27, 323–338 (2019).
Ozdemir, T., Fedorec, A. J. H., Danino, T. & Barnes, C. P. Synthetic biology and engineered live biotherapeutics: toward increasing system complexity. Cell Syst. 7, 5–16 (2018).
Fehér, C., Soriano, A. & Mensa, J. A review of experimental and off-label therapies for clostridium difficile infection. Infect. Dis. Ther. 6, 1–35 (2017).
Connelly, S., Fanelli, B., Hasan, N. A., Colwell, R. R. & Kaleko, M. Oral metallo-beta-lactamase protects the gut microbiome from carbapenem-mediated damage and reduces propagation of antibiotic resistance in pigs. Front Microbiol. 10, 101–101 (2019).
Ramachandran, G. & Bikard, D. Editing the microbiome the CRISPR way. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 374, 20180103 (2019). New technologies bring challenges and opportunities for future new therapies that are reviewed in this article.
Pena, R. T. et al. Relationship between quorum sensing and secretion systems. Front Microbiol. 10, 1100–1100 (2019).
Sharma, D., Misba, L. & Khan, A. U. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 8, 76–76 (2019).
Murugayah, S. A. & Gerth, M. L. Engineering quorum quenching enzymes: progress and perspectives. Biochem. Soc. Trans. 47, 793–800 (2019).
Defraine, V., Fauvart, M. & Michiels, J. Fighting bacterial persistence: Current and emerging anti-persister strategies and therapeutics. Drug Resist. Updat. 38, 12–26 (2018).
Fleitas Martínez, O., Cardoso, M. H., Ribeiro, S. M. & Franco, O. L. Recent advances in anti-virulence therapeutic strategies with a focus on dismantling bacterial membrane microdomains, toxin neutralization, quorum-sensing interference and biofilm inhibition. Front Cell Infect. Microbiol. 9, 74–74 (2019).
Maura, D., Ballok, A. E. & Rahme, L. G. Considerations and caveats in anti-virulence drug development. Curr. Opin. Microbiol. 33, 41–46 (2016).
Dickey, S. W., Cheung, G. Y. C. & Otto, M. Different drugs for bad bugs: antivirulence strategies in the age of antibiotic resistance. Nat. Rev. Drug Discov. 16, 457–471 (2017).
Aridis Pharmaceuticals, Inc. Aridis Pharmaceuticals reports phase 2 clinical trial results of AR-105 for the treatment of ventilator-associated pneumonia caused by Pseudomonas Aeruginosa. Investors.aridispharma.com. https://investors.aridispharma.com/2019-09-03-Aridis-Pharmaceuticals-Reports-Phase-2-Clinical-Trial-Results-of-AR-105-for-the-Treatment-of-Ventilator-Associated-Pneumonia-Caused-by-Pseudomonas-Aeruginosa (2019).
Redi, D., Raffaelli, C. S., Rossetti, B., De Luca, A. & Montagnani, F. Staphylococcus aureus vaccine preclinical and clinical development: current state of the art. New Microbiol. 41, 208–213 (2018).
Priebe, G. P. & Goldberg, J. B. Vaccines for pseudomonas aeruginosa: a long and winding road. Expert Rev. Vaccines 13, 507–519 (2014).
Canaparo, R. et al. Recent developments in antibacterial therapy: focus on stimuli-responsive drug-delivery systems and therapeutic nanoparticles. Molecules 24, 1991 (2019).
Kwon, E. J. et al. Porous silicon nanoparticle delivery of tandem peptide anti-infectives for the treatment of pseudomonas aeruginosa lung infections. Adv. Mater. 29, 1701527 (2017).
Kumar, M., Curtis, A. & Hoskins, C. Application of nanoparticle technologies in the combat against anti-microbial resistance. Pharmaceutics. 10, 11 (2018).
Theuretzbacher, U. & Paul, M. Developing a new antibiotic for extensively drug-resistant pathogens: the case of plazomicin. Clin. Microbiol. Inf. 24, 1231–1233 (2018).
Czaplewski, L. et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 16, 239–251 (2016).
Acknowledgements
U.T. received federal funds from the German Federal Ministry of Education and Research (BMBF) to support this Review. The funder had no role in study design, data collection, data analysis, data interpretation or writing of the article. K.O. is Principal Investigator and Executive Director of CARB-X, funded by Notice of Award 5IDSEP160030-04-00 from the US Biomedical Advanced Research and Development Authority (Assistant Secretary for Preparedness and Response, Department of Health and Human Services), preclinical services from the US National Institute for Allergy and Infectious Diseases (National Institutes of Health, Department of Health and Human Services) and funding from the Wellcome Trust, the German Federal Ministry for Education and Research (BMBF), the Global AMR Innovation Fund from the UK Department of Health and Social Care and the Bill & Melinda Gates Foundation. A.E. is a partner with Novo Holdings A/S and Director of the REPAIR Impact Fund, which was commissioned by the Novo Nordisk Foundation. A.K. is the leader of the managing entity of the ENABLE consortium and has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115583, resources of which are composed of financial contributions from the European Union’s seventh framework programme and European Federation of Pharmaceutical Industries and Associations companies’ in-kind contribution. Those projects in ENABLE that volunteered information for this Review are gratefully acknowledged. The views herein do not necessarily represent the view of the ENABLE partners. The views expressed herein do not necessarily represent the views of CARB-X or any CARB-X funder, the REPAIR Impact Fund or ENABLE or any of its funders. The authors thank L. Marin for providing data from the Joint Programming Initiative on Antimicrobial Resistance.
Author information
Authors and Affiliations
Contributions
A.E. provided the data from the REPAIR Impact Fund. K.O. provided the data from CARB-X. A.K. provided the data from ENABLE. U.T. provided the data from the Center for Anti-Infective Agents (CEFAIA). All data was provided to U.T. for application of the inclusion criteria, descriptive results, analysis and discussion of the findings. U.T. wrote the first draft and K.O. provided the first comprehensive edit, which was then reviewed and edited by all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
CARB-X: https://carb-x.org/portfolio/gallery/
ENABLE: http://nd4bb-enable.eu/enable-portfolio
Joint Programming Initiative on Antimicrobial Resistance: https://www.jpiamr.eu/supportedprojects/
REPAIR Impact Fund: https://www.repair-impact-fund.com/portfolio/
Supplementary information
Glossary
- Lead generation phase (hit-to-lead phase)
-
Drug discovery phase where promising molecules (hits) are evaluated and undergo limited optimization to identify suitable lead compounds.
- Repurposed approved drugs
-
Repurposing a drug is a strategy for identifying new uses for an approved drug that are outside the scope of the original indication.
- Label expansion
-
Aims to achieve additional regulatory approval for a new indication beyond the original use for which the drug was approved.
- Indications
-
A therapeutic indication refers to the use of a drug for treating a particular disease. The indication can be approved by regulatory agencies or not approved.
- Spectrum
-
Range of activity against a group of bacteria.
- Formulations
-
Pharmaceutical formulation is the process in which the active compound and additional ingredients are combined to produce a final medicinal product.
- Parenteral application
-
Route of administration other than the gastrointestinal tract to achieve systemic distribution.
- Intravesical application
-
Administration of a drug directly into the bladder.
- Non-fermenters
-
Heterogeneous group of bacteria which cannot use glucose and thus are unable to generate energy through fermentation of glucose. Important genera of non-fermenters include Pseudomonas and Acinetobacter.
- Phase III trials
-
In clinical development, phase III clinical trials are randomized controlled multicentre studies that assess the effectiveness and safety of a drug in comparison to current standard-of-care treatment.
- Metallo-ß-lactamases
-
ß-Lactamases that require zinc for activity and hydrolyse penicillins, cephalosporins and carbapenems.
- Endolysins
-
Enzymes that are produced by bacteriophages and hydrolyse the bacterial cell wall to escape the cell at the end of the cycle.
- Compassionate use
-
The use of unapproved drugs outside clinical trials for patients without options of treatment with an approved drug.
- Push funding
-
Incentivizes discovery and development activities before achieving regulatory approval.
- Pull incentives
-
Reward the successful development and regulatory approval of a new drug.
Rights and permissions
About this article
Cite this article
Theuretzbacher, U., Outterson, K., Engel, A. et al. The global preclinical antibacterial pipeline. Nat Rev Microbiol 18, 275–285 (2020). https://doi.org/10.1038/s41579-019-0288-0
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41579-019-0288-0
This article is cited by
-
Natural flavonoids combat cytosolic MRSA by potentiating phagosome acidification
Cell Communication and Signaling (2025)
-
Targeting the G-quadruplex as a novel strategy for developing antibiotics against hypervirulent drug-resistant Staphylococcus aureus
Journal of Biomedical Science (2025)
-
Antibiotics re-booted—time to kick back against drug resistance
npj Antimicrobials and Resistance (2025)
-
Global health perspectives on antibacterial drug discovery and the preclinical pipeline
Nature Reviews Microbiology (2025)
-
Design, synthesis and optimization of TarO inhibitors as multifunctional antibiotics against Methicillin-resistant Staphylococcus aureus
npj Antimicrobials and Resistance (2025)