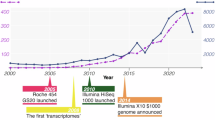
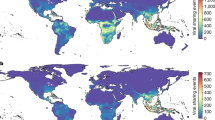
Abstract
The COVID-19 pandemic has given the study of virus evolution and ecology new relevance. Although viruses were first identified more than a century ago, we likely know less about their diversity than that of any other biological entity. Most documented animal viruses have been sampled from just two phyla — the Chordata and the Arthropoda — with a strong bias towards viruses that infect humans or animals of economic and social importance, often in association with strong disease phenotypes. Fortunately, the recent development of unbiased metagenomic next-generation sequencing is providing a richer view of the animal virome and shedding new light on virus evolution. In this Review, we explore our changing understanding of the diversity, composition and evolution of the animal virome. We outline the factors that determine the phylogenetic diversity and genomic structure of animal viruses on evolutionary timescales and show how this impacts assessment of the risk of disease emergence in the short term. We also describe the ongoing challenges in metagenomic analysis and outline key themes for future research. A central question is how major events in the evolutionary history of animals, such as the origin of the vertebrates and periodic mass extinction events, have shaped the diversity and evolution of the viruses they carry.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Wasik, B. R. & Turner, P. E. On the biological success of viruses. Annu. Rev. Microbiol. 67, 519–541 (2013).
Holmes, E. C. The Evolution and Emergence of RNA Viruses (Oxford University Press, 2009).
Loeffler, F. A. J. & Frosch, P. Berichte der Kommission zur Erforschung der Maul- und Klauenseuche bei dem Institut für Infektionskrankheiten in Berlin (G. Fischer, 1898).
Kumar, A., Murthy, S. & Kapoor, A. Evolution of selective-sequencing approaches for virus discovery and virome analysis. Virus Res. 239, 172–179 (2017).
Shi, M. et al. Redefining the invertebrate RNA virosphere. Nature 540, 539–543 (2016).
Shi, M. et al. Divergent viruses discovered in arthropods and vertebrates revise the evolutionary history of the Flaviviridae and related viruses. J. Virol. 90, 659–669 (2015).
Zhang, Y. Z., Shi, M. & Holmes, E. C. Using metagenomics to characterize an expanding virosphere. Cell 172, 1168–1172 (2018).
Ambrose, H. E. & Clewley, J. P. Virus discovery by sequence-independent genome amplification. Rev. Med. Virol. 16, 365–383 (2006).
Shi, M. et al. The evolutionary history of vertebrate RNA viruses. Nature 556, 197–202 (2018). Major study of the phylogenetic diversity of RNA viruses carried by diverse vertebrates, showing that many of the virus families associated with mammals have a deep ancestry with evolutionary roots in fish.
Buchfink, B., Reuter, K. & Drost, H. G. Sensitive protein alignments at tree-of-life scale using diamond. Nat. Methods 18, 366–368 (2021).
Geoghegan, J. L. & Holmes, E. C. Predicting virus emergence amid evolutionary noise. Open Biol. 7, 170189 (2017).
Fernández, R. & Gabaldón, T. Gene gain and loss across the metazoan tree of life. Nat. Ecol. Evol. 4, 524–533 (2020).
Laumer, C. E. et al. Revisiting metazoan phylogeny with genomic sampling of all phyla. Proc. Biol. Sci. 286, 20190831 (2019).
Paraskevopoulou, S. et al. Viromics of extant insect orders unveil the evolution of the flavi-like superfamily. Virus Evol. 7, veab030 (2021).
Murphy, F. A. Historical perspective: what constitutes discovery (of a new virus)? Adv. Virus Res. 95, 197–220 (2016).
Greninger, A. L. A decade of RNA virus metagenomics is (not) enough. Virus Res. 244, 218–229 (2018).
Li, C. X. et al. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. eLife 4, e05378 (2015). First article to show that invertebrates — in this case arthropods — harbour an enormous diversity of RNA viruses, often at high abundance. Provides the first description of the chuviruses, which are characterized by diverse genome structures.
Donaldson, E. F. et al. Metagenomic analysis of the viromes of three North American bat species: viral diversity among different bat species that share a common habitat. J. Virol. 84, 13004–13018 (2010).
Li, L. et al. The fecal viral flora of California sea lions. J. Virol. 85, 9909–9917 (2011).
Ge, X. et al. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 86, 4620–4630 (2012).
Walker, P. J. et al. Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch. Virol. https://doi.org/10.1007/s00705-021-05156-1 (2021).
Lauber, C. et al. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe 22, 387–399.e386 (2017). Major study of the phylogenetic diversity of HBV-like viruses in fish, including the discovery of a group of related viruses — the nackednaviruses — that lack the envelope protein.
Geoghegan, J. L. et al. Hidden diversity and evolution of viruses in market fish. Virus Evol. 4, vey031 (2018).
Zeigler Allen, L. et al. The Baltic Sea virome: diversity and transcriptional activity of DNA and RNA viruses. mSystems 2, e00125–16 (2017).
Parry, R., Wille, M., Turnbull, O. M. H., Geoghegan, J. L. & Holmes, E. C. Divergent influenza-like viruses of amphibians and fish support an ancient evolutionary association. Viruses 12, 1042 (2020).
Geoghegan, J. L. et al. Virome composition in marine fish revealed by meta-transcriptomics. Virus Evol. 7, veab005 (2021).
Costa, V. A. et al. Metagenomic sequencing reveals a lack of virus exchange between native and invasive freshwater fish across the Murray–Darling Basin, Australia. Virus Evol. 7, veab034 (2021).
Miller, A. K. et al. Slippery when wet: cross-species transmission of divergent coronaviruses in bony and jawless fish and the evolutionary history of the Coronaviridae. Virus Evol. 7, veab050 (2021).
López-Bueno, A. et al. Concurrence of iridovirus, polyomavirus, and a unique member of a new group of fish papillomaviruses in lymphocystis disease-affected gilthead sea bream. J. Virol. 90, 8768–8779 (2016).
Wu, F. et al. A new coronavirus associated with human respiratory disease in China. Nature 579, 265–269 (2020).
Lu, R. et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395, 565–574 (2020).
Wang, W. et al. Extensive genetic diversity and host range of rodent-borne coronaviruses. Virus Evol. 6, veaa078 (2021).
Latinne, A. et al. Origin and cross-species transmission of bat coronaviruses in China. Nat. Commun. 11, 4235 (2020).
Temmam, S. et al. Coronaviruses with a SARS-CoV-2-like receptor-binding domain allowing ACE2-mediated entry into human cells isolated from bats of Indochinese peninsula. Preprint at Research Square https://doi.org/10.21203/rs.3.rs-871965/v1 (2021).
van Aart, A. E. et al. SARS-CoV-2 infection in cats and dogs in infected mink farms. Transbound. Emerg. Dis. https://doi.org/10.1111/tbed.14173 (2021).
Oude Munnink, B. B. et al. Transmission of SARS-CoV-2 on mink farms between humans and mink and back to humans. Science 371, 172–177 (2021). Demonstration of the broad host range of SARS-CoV-2, reflected in a major outbreak in farmed mink. That the virus was able to spread back to humans shows that some animal species may become SARS-CoV-2 reservoirs.
Chandler, J. C. et al. SARS-CoV-2 exposure in wild white-tailed deer (Odocoileus virginianus). Preprint at bioRxiv https://doi.org/10.1101/2021.07.29.454326 (2021).
Lam, T. T.-Y. et al. Identifying SARS-CoV-2 related coronaviruses in Malayan pangolins. Nature 583, 282–285 (2020).
Zhou, H. et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell 184, 4380–4391 (2021).
Corman, V. M., Muth, D., Niemeyer, D. & Drosten, C. Hosts and sources of endemic human coronaviruses. Adv. Virus Res. 100, 163–188 (2018). Important review of the human coronaviruses highlighting their diversity, evolutionary history and zoonotic origins.
Edgar, R. C. et al. Petabase-scale sequence alignment catalyses viral discovery. Preprint at bioRxiv https://doi.org/10.1101/2020.08.07.241729 (2020).
Salehi-Ashtiani, K., Lupták, A., Litovchick, A. & Szostak, J. W. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science 313, 1788–1792 (2006).
Chang, W.-S. et al. Novel hepatitis D-like agents in vertebrates and invertebrates. Virus Evol. 5, vez021 (2019).
Hetzel, U. et al. Identification of a novel deltavirus in boa constrictors. mBio 10, e00014-19 (2019).
Iwamoto, M. et al. Identification of novel avian and mammalian deltaviruses provides new insights into deltavirus evolution. Virus Evol. 7, veab003 (2021). Overview of the evolution of deltaviruses (that is, HDV-like viruses) in birds and mammals. Reveals the ancient history and diversity of these viruses and shows that they are not exclusively associated with humans or HBV.
Paraskevopoulou, S. et al. Mammalian deltavirus without hepadnavirus coinfection in the neotropical rodent Proechimys semispinosus. Proc. Natl Acad. Sci. USA 117, 17977–17983 (2020).
Taubenberger, J. K. & Kash, J. C. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7, 440–451 (2010).
Joseph, U., Su, Y. C., Vijaykrishna, D. & Smith, G. J. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respir. Viruses 11, 74–84 (2017).
Wu, H. et al. Abundant and diverse RNA viruses in insects revealed by RNA-Seq analysis: ecological and evolutionary implications. mSystems 5, e00039-20 (2020).
Van Eynde, B. et al. Exploration of the virome of the European brown shrimp (Crangon crangon). J. Gen. Virol. 101, 651–666 (2020).
Laffy, P. W. et al. Reef invertebrate viromics: diversity, host specificity and functional capacity. Environ. Microbiol. 20, 2125–2141 (2018).
Tokarz, R. et al. Virome analysis of Amblyomma americanum, Dermacentor variabilis, and Ixodes scapularis ticks reveals novel highly divergent vertebrate and invertebrate viruses. J. Virol. 88, 11480–11492 (2014).
Käfer, S. et al. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog. 15, e1008224 (2019).
Ramírez, A. L. et al. Metagenomic analysis of the virome of mosquito excreta. mSphere 5, e00587-20 (2020).
Brinkmann, A. et al. A metagenomic survey identifies Tamdy orthonairovirus as well as divergent phlebo-, rhabdo-, chu- and flavi-like viruses in Anatolia, Turkey. Ticks Tick. Borne Dis. 9, 1173–1183 (2018).
Gudenkauf, B. M. & Hewson, I. Comparative metagenomics of viral assemblages inhabiting four phyla of marine invertebrates. Front. Mar. Sci. 3, 23 (2016).
Medd, N. C. et al. The virome of Drosophila suzukii, an invasive pest of soft fruit. Virus Evol. 4, vey009 (2018).
Webster, C. L. et al. The discovery, distribution, and evolution of viruses associated with Drosophila melanogaster. PLoS Biol. 13, e1002210 (2015).
Hameed, M. et al. A metagenomic analysis of mosquito virome collected from different animal farms at Yunnan–Myanmar border of China. Front. Microbiol. 11, 591478 (2021).
Sadeghi, M. et al. Virome of >12 thousand Culex mosquitoes from throughout California. Virology 523, 74–88 (2018). Major metagenomic study of virome diversity in mosquitoes showing the power of this technology for high-throughput virus screening at a single location.
He, X. et al. Metagenomic sequencing reveals viral abundance and diversity in mosquitoes from the Shaanxi-Gansu-Ningxia region, China. PLoS Negl. Trop. Dis. 15, e0009381 (2021).
Marklewitz, M., Zirkel, F., Kurth, A., Drosten, C. & Junglen, S. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc. Natl Acad. Sci. USA 112, 7536–7541 (2015).
Schmidlin, K. et al. A novel lineage of polyomaviruses identified in bark scorpions. Virology 563, 58–63 (2021).
Qin, X. C. et al. A tick-borne segmented RNA virus contains genome segments derived from unsegmented viral ancestors. Proc. Natl Acad. Sci. USA 111, 6744–6749 (2014).
Ladner, J. T. et al. A multicomponent animal virus isolated from mosquitoes. Cell Host Microbe 20, 357–367 (2016). First description of a multicomponent virus in an animal. Highlights the complexity of genome evolution in RNA viruses, in this case in the flavi-like viruses.
Argenta, F. F. et al. Identification of reptarenaviruses, hartmaniviruses, and a novel chuvirus in captive native Brazilian boa constrictors with boid inclusion body disease. J. Virol. 94, e00001-20 (2020).
Sharp, P. M. & Hahn, B. H. Origins of HIV and the AIDS pandemic. Cold Spring Harb. Perspect. Med. 1, a006841 (2011).
Pybus, O. G., Rambaut, A., Holmes, E. C. & Harvey, P. H. New inferences from tree shape: numbers of missing taxa and population growth rates. Syst. Biol. 51, 881–888 (2002).
Kapusinszky, B. et al. Local virus extinctions following a host population bottleneck. J. Virol. 89, 8152–8161 (2015).
McKee, C. D., Bai, Y., Webb, C. T. & Kosoy, M. Y. Bats are key hosts in the radiation of mammal-associated Bartonella bacteria. Infect. Genet. Evol. 89, 104719 (2021).
dos Reis, M. et al. Uncertainty in the timing of origin of animals and the limits of precision in molecular timescales. Curr. Biol. 25, 2939–2950 (2015).
Flajnik, M. F. & Kasahara, M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 11, 47–59 (2010).
Wang, L.-F., Walker, P. J. & Poon, L. L. M. Mass extinctions, biodiversity and mitochondrial function: are bats ‘special’ as reservoirs for emerging viruses? Curr. Opin. Virol. 1, 649–657 (2011). One of the first articles to propose that bats are uniquely important hosts for emerging viruses and that host mass extinction events might play a key role in shaping the phylogenetic diversity of viruses.
Stanley, S. M. Estimates of the magnitudes of major marine mass extinctions in earth history. Proc. Natl Acad. Sci. USA 113, E6325–E6334 (2016).
Raup, D. M. & Sepkoski, J. J. Mass extinctions in the marine fossil record. Science 215, 1501–1503 (1982).
Lynch, M. & Conery, J. S. The origins of genome complexity. Science 302, 1401–1404 (2003).
Carlson, C. J., Zipfel, C. M., Garnier, R. & Bansal, S. Global estimates of mammalian viral diversity accounting for host sharing. Nat. Ecol. Evol. 3, 1070–1075 (2019).
Geoghegan, J. L., Duchêne, S. & Holmes, E. C. Comparative analysis estimates the relative frequencies of co-divergence and cross-species transmission within viral families. PLoS Pathog. 13, e1006215 (2017).
Racaniello, V. Moving beyond metagenomics to find the next pandemic virus. Proc. Natl Acad. Sci. USA 113, 2812–2814 (2016).
Morse, S. S. et al. Prediction and prevention of the next pandemic zoonosis. Lancet 380, 1956–1965 (2012).
Smith, I. & Wang, L. F. Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 3, 84–91 (2013).
Olival, K. J. et al. Host and viral traits predict zoonotic spillover from mammals. Nature 546, 646–650 (2017).
Wasik, B. R. et al. Onward transmission of viruses: how do viruses emerge to cause epidemics after spillover? Philos. Trans. R. Soc. Lond. B Biol. Sci. 374, 20190017 (2019).
Düx, A. et al. Measles virus and rinderpest virus divergence dated to the sixth century BCE. Science 368, 1367–1370 (2020).
Tabachnick, W. J. Climate change and the arboviruses: lessons from the evolution of the dengue and yellow fever viruses. Annu. Rev. Virol. 3, 125–145 (2016).
Fritzell, C. et al. Current challenges and implications for dengue, chikungunya and Zika seroprevalence studies worldwide: a scoping review. PLoS Negl. Trop. Dis. 12, e0006533 (2018).
Campbell-Lendrum, D., Manga, L., Bagayoko, M. & Sommerfeld, J. Climate change and vector-borne diseases: what are the implications for public health research and policy? Philos. Trans. R. Soc. Lond. B. Biol. Sci. 370, 20130552 (2015).
Jacob, S. T. et al. Ebola virus disease. Nat. Rev. Dis. Prim. 6, 13 (2020).
Gould, E. A. & Higgs, S. Impact of climate change and other factors on emerging arbovirus diseases. Trans. R. Soc. Trop. Med. Hyg. 103, 109–121 (2009).
Marcondes, M. & Day, M. J. Current status and management of canine leishmaniasis in Latin America. Res. Vet. Sci. 123, 261–272 (2019).
Brock, P. M. et al. Predictive analysis across spatial scales links zoonotic malaria to deforestation. Proc. Biol. Sci. 286, 20182351 (2019).
Ayala, A. J., Yabsley, M. J. & Hernandez, S. M. A review of pathogen transmission at the backyard chicken–wild bird interface. Front. Vet. Sci. 7, 662 (2020).
Munoz, O. et al. Genetic adaptation of influenza A viruses in domestic animals and their potential role in interspecies transmission: a literature review. Ecohealth 13, 171–198 (2016).
Peiris, J. S., de Jong, M. D. & Guan, Y. Avian influenza virus (H5N1): a threat to human health. Clin. Microbiol. Rev. 20, 243–267 (2007). Review of the ecology and evolution of H5N1 avian influenza virus, particularly how it emerges in humans from its avian reservoir populations and its associated pandemic risk.
Schelling, E., Thur, B., Griot, C. & Audige, L. Epidemiological study of Newcastle disease in backyard poultry and wild bird populations in Switzerland. Avian Pathol. 28, 263–272 (1999).
Boros, Á. et al. A diarrheic chicken simultaneously co-infected with multiple picornaviruses: complete genome analysis of avian picornaviruses representing up to six genera. Virology 489, 63–74 (2016).
Lang, A. S. et al. Assessing the role of seabirds in the ecology of influenza A viruses. Avian Dis. 60, 378–386 (2016).
Lickfett, T. M., Clark, E., Gehring, T. M. & Alm, E. W. Detection of influenza A viruses at migratory bird stopover sites in Michigan, USA. Infect. Ecol. Epidemiol. 8, 1474709 (2018).
Rezza, G. Dengue and chikungunya: long-distance spread and outbreaks in naïve areas. Pathog. Glob. Health 108, 349–355 (2014).
Fritzsche McKay, A. & Hoye, B. J. Are migratory animals superspreaders of infection? Integr. Comp. Biol. 56, 260–267 (2016).
Jeong, S. et al. Introduction of avian influenza A(H6N5) virus into Asia from North America by wild birds. Emerg. Infect. Dis. 25, 2138–2140 (2019).
Pettersson, J. H. O. et al. Circumpolar diversification of the Ixodes uriae tick virome. PLoS Pathog. 16, e1008759 (2020).
Albery, G. F., Eskew, E. A., Ross, N. & Olival, K. J. Predicting the global mammalian viral sharing network using phylogeography. Nat. Commun. 11, 2260 (2020).
Dill, J. A. et al. Distinct viral lineages from fish and amphibians reveal the complex evolutionary history of hepadnaviruses. J. Virol. 90, 7920–7933 (2016).
Mollentze, N. & Streicker, D. G. Viral zoonotic risk is homogenous among taxonomic orders of mammalian and avian reservoir hosts. Proc. Natl Acad. Sci. USA 117, 9423–9430 (2020).
Mollentze, N., Babayan, S. A. & Streicker, D. G. Identifying and prioritizing potential human-infecting viruses from their genome sequences. PLoS Biol. 19, e3001390 (2021).
Wille, M., Geoghegan, J. L. & Holmes, E. C. How accurately can we assess zoonotic risk? PLoS Biol. 19, e3001135 (2021).
Kohl, C. et al. The virome of German bats: comparing virus discovery approaches. Sci. Rep. 11, 7430 (2021).
Li, L. et al. Bat guano virome: predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 84, 6955–6965 (2010).
Kemenesi, G. et al. Molecular survey of RNA viruses in Hungarian bats: discovering novel astroviruses, coronaviruses, and caliciviruses. Vector Borne Zoonotic Dis. 14, 846–855 (2014).
Letko, M., Seifert, S. N., Olival, K. J., Plowright, R. K. & Munster, V. J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 18, 461–471 (2020). Extensive review of the relevant biology of bats and the viruses they carry, particularly in the context of SARS-CoV-2.
Irving, A. T., Ahn, M., Goh, G., Anderson, D. E. & Wang, L.-F. Lessons from the host defences of bats, a unique viral reservoir. Nature 589, 363–370 (2021). Timely review outlining the reasons why bats might be uniquely important virus reservoirs and what this might mean for understanding future emergence events.
Banerjee, A. et al. Novel insights into immune systems of bats. Front. Immunol. 11, 26 (2020).
Holmes, E. C., Rambaut, A. & Andersen, K. G. Pandemics: spend on surveillance, not prediction. Nature 558, 180–182 (2018).
Sanjuán, R. & Domingo-Calap, P. Mechanisms of viral mutation. Cell Mol. Life Sci. 73, 4433–4448 (2016).
Plowright, R. K. et al. Pathways to zoonotic spillover. Nat. Rev. Microbiol. 15, 502–510 (2017). Benchmark review of the ecological processes by which viruses can spill over and emerge in new hosts, identifying this as a key process in virus evolution.
Holmes, E. C. et al. The origins of SARS-CoV-2: a critical review. Cell 184, 4848–4856 (2021).
Johnson, B. A. et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature 591, 293–299 (2021).
Wrobel, A. G. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 27, 763–767 (2020). Detailed structural virology study that demonstrates that even closely related human and animal coronaviruses can differ profoundly in receptor-binding ability.
Wilson, M. R. et al. Chronic meningitis investigated via metagenomic next-generation sequencing. JAMA Neurol. 75, 947–955 (2018).
Wilson, M. R. et al. Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N. Engl. J. Med. 380, 2327–2340 (2019). Key article showing the importance of mNGS in a clinical diagnostic setting, in this case for the identification of the microbial pathogens associated with meningitis and encephalitis.
Xu, G. J. et al. Comprehensive serological profiling of human populations using a synthetic human virome. Science 348, aaa0698 (2015). Presents VirScan — a method for the high-throughput screening of viruses by identifying antiviral antibodies in human sera. Although originally designed to screen the human virome, the method could be adapted to detect zoonotic viruses.
Field, H. E., Mackenzie, J. S. & Daszak, P. Henipaviruses: emerging paramyxoviruses associated with fruit bats. Curr. Top. Microbiol. Immunol. 315, 133–159 (2007).
Harvey, E. et al. Extensive diversity of RNA viruses in Australian ticks. J. Virol. 93, e01358-18 (2019).
Wille, M. et al. Sustained RNA virome diversity in Antarctic penguins and their ticks. ISME J. 14, 1768–1782 (2020).
Di Giallonardo, F., Schlub, T. E., Shi, M. & Holmes, E. C. Dinucleotide composition in RNA viruses is shaped more by virus family than host species. J. Virol. 91, e02381-16 (2017).
Krishnamurthy, S. R. & Wang, D. Origins and challenges of viral dark matter. Virus Res. 239, 136–142 (2017).
Bamford, D. H., Grimes, J. M. & Stuart, D. I. What does structure tell us about virus evolution? Curr. Opin. Struct. Biol. 15, 655–663 (2005). Demonstrates how patterns of evolutionary relatedness are preserved in the structure of viral capsid proteins. Lays the foundation for how protein structural information can be used to infer phylogenetic relationships.
Illergård, K., Ardell, D. H. & Elofsson, A. Structure is three to ten times more conserved than sequence — a study of structural response in protein cores. Proteins 77, 499–508 (2009).
Harrison, S. C. Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 (2008).
Fédry, J. et al. The ancient gamete fusogen HAP2 is a eukaryotic class II fusion protein. Cell 168, 904–915 (2017). Demonstration of how protein structure can reveal ancient evolutionary homologies, in this case between an algal gamete fusogen and a class II viral membrane fusion protein.
Henderson, R. Overview and future of single particle electron cryomicroscopy. Arch. Biochem. Biophys. 581, 19–24 (2015).
Eddy, S. R. Accelerated profile HMM searches. PLoS Comp. Biol. 7, e1002195 (2011).
Krupovic, M., Cvirkaite-Krupovic, V., Iranzo, J., Prangishvili, D. & Koonin, E. V. Viruses of Archaea: structural, functional, environmental and evolutionary genomics. Virus Res. 244, 181–193 (2018).
Holmes, E. C. & Duchêne, S. Can sequence phylogenies safely infer the origin of the global virome? mBio 10, e00289-19 (2019).
Chang, G. S. et al. Phylogenetic profiles reveal evolutionary relationships within the ‘twilight zone’ of sequence similarity. Proc. Natl Acad. Sci. USA 105, 13474–13479 (2008).
Nguyen, L.-T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Wooley, J. C., Godzik, A. & Friedberg, I. A primer on metagenomics. PLoS Comp. Biol. 6, e10006677 (2010).
O’Neil, D., Glowatz, H. & Schlumpberger, M. Ribosomal RNA depletion for efficient use of RNA-seq capacity. Curr. Protoc. Mol. Biol. https://doi.org/10.1002/0471142727.mb0419s103 (2013).
Briese, T. et al. Virome capture sequencing enables sensitive viral diagnosis and comprehensive virome analysis. mBio 6, e01491-15 (2015).
Chong, R. et al. Fecal viral diversity of captive and wild Tasmanian devils characterized using virion-enriched metagenomics and metatranscriptomics. J. Virol. 93, e00205-19 (2019).
Author information
Authors and Affiliations
Contributions
E.H. researched data for the article. Both authors contributed substantially to discussion of the content, wrote the article, and edited and reviewed the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Microbiology thanks Kevin Olival, Arvind Varsani and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Metagenomic next-generation sequencing
-
(mNGS). The parallel high-throughput sequencing of the total genetic material (RNA or DNA) extracted from a sample. This method offers scalability and speed that cannot be achieved by earlier sequencing technologies.
- Virosphere
-
The total assemblage of RNA viruses and DNA viruses on Earth, infecting hosts of any type.
- Viromes
-
Total assemblages of viruses in individual organisms or species.
- Metatranscriptomics
-
The study of the total expressed RNA — the transcriptome — within a sample. The RNA can be derived from expressed host genes as well as microbial species within the host, including both RNA virsuses and DNA viruses.
- Zoonotic disease
-
An infectious disease that can be transmitted from animals to humans.
- Emergence
-
Process by which novel infectious diseases (or pathogens) appear in species or previously known diseases rapidly increase in incidence or geographical range. Often associated with cross-species transmission.
- Metagenomics
-
The simultaneous sequencing of all genetic material within a sample, including all the microorganisms present. It can involve the analysis of individual marker genes such as 16S or 18S ribosomal RNA or complete genomes.
- Co-divergence
-
Evolutionary pattern in which the phylogenetic history of a virus or other pathogen matches that of the host organisms on long evolutionary timescales.
- Multicomponent viruses
-
Also referred to ‘multipartite viruses’. Viruses in which the genome segments are contained within separate virus particles. These are relatively commonplace in positive-sense RNA viruses of plants such as members of the Bromoviridae.
- Genetic drift
-
The change in frequency of a mutation in a population due to the chance effect of random sampling. Although genetic drift occurs in all populations of finite size, its effect is strongest in small populations.
- Cross-species transmission
-
Also referred to as ‘host-jumping’ or ‘host-switching’. The transmission of a virus from one host species to another.
- Ectoparasites
-
Parasitic organisms that live on the skin of the host (rather than within a host), from which they derive their energy.
- Spillover
-
The initial and sometimes transient appearance of a pathogen in a new species following a host jump. Can sometimes lead to a full-blown epidemic or pandemic.
Rights and permissions
About this article
Cite this article
Harvey, E., Holmes, E.C. Diversity and evolution of the animal virome. Nat Rev Microbiol 20, 321–334 (2022). https://doi.org/10.1038/s41579-021-00665-x
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41579-021-00665-x
This article is cited by
-
The genomic diversity of arthropod-specific viruses reinforces the continental distribution pattern of Aedes aegypti
Parasites & Vectors (2025)
-
Host ecology and phylogeny shape the temporal dynamics of social bee viromes
Nature Communications (2025)
-
MALDI-TOF MS technique as a new approach for simultaneous detection and differentiation of potato virus Y strains
Scientific Reports (2025)
-
Node role of wild boars in virus circulation among wildlife and domestic animals
Nature Communications (2025)
-
Evaluation of Enrichment Approaches for the Study of the Viromes in Mollusk Species
Food and Environmental Virology (2025)