Abstract
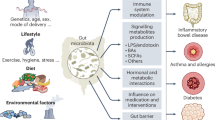
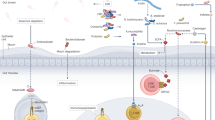
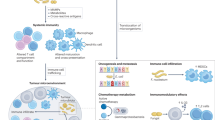
The microbiome is increasingly recognized as a key player in cancer pathogenesis and treatment response, acting through both local and systemic mechanisms. Microbial communities and their metabolites can directly influence drug metabolism, shape the immune landscape, and alter transcriptional and epigenetic programmes in the gut, systemically and in the tumour microenvironment. Emerging data support the potential of microbiome-targeted interventions (such as faecal microbiota transplantation, diet, prebiotics and probiotics) as adjuncts to conventional cancer therapies, with the goal of enhancing efficacy and reducing toxicity. This Review highlights the promise of the microbiome as a prognostic and predictive biomarker, a modifiable factor in cancer care and prevention, and a therapeutic target. We also discuss major knowledge gaps, limitations in current study designs, and the need for mechanism-guided, personalized strategies to advance clinical translation.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
McCallum, G. & Tropini, C. The gut microbiota and its biogeography. Nat. Rev. Microbiol. 22, 105–118 (2024).
Verdegaal, A. A. & Goodman, A. L. Integrating the gut microbiome and pharmacology. Sci. Transl. Med. 16, eadg8357 (2024).
Yang, L., Li, A., Wang, Y. & Zhang, Y. Intratumoral microbiota: roles in cancer initiation, development and therapeutic efficacy. Signal Transduct. Target. Ther. 8, 35 (2023).
Domzaridou, E. et al. The impact of oral antibiotics prior to cancer diagnosis on overall patient survival: findings from an English population-based cohort study. Curr. Oncol. 30, 8434–8443 (2023).
Yuan, L. et al. The influence of gut microbiota dysbiosis to the efficacy of 5-fluorouracil treatment on colorectal cancer. Biomed. Pharmacother. 108, 184–193 (2018).
Routy, B. et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). This groundbreaking translational study demonstrates that the microbiome can enhance the efficacy of ICIs by modulating antitumour immune responses in both human cohorts and animal models.
Elkrief, A. et al. Antibiotics are associated with worse outcomes in lung cancer patients treated with chemotherapy and immunotherapy. NPJ Precis. Oncol. 8, 143 (2024).
Ransohoff, J. D. et al. Antimicrobial exposure is associated with decreased survival in triple-negative breast cancer. Nat. Commun. 14, 2053 (2023).
Cong, J. et al. Bile acids modified by the intestinal microbiota promote colorectal cancer growth by suppressing CD8+ T cell effector functions. Immunity 57, 876–889.e11 (2024).
Tintelnot, J. et al. Microbiota-derived 3-IAA influences chemotherapy efficacy in pancreatic cancer. Nature 615, 168–174 (2023). Using human cohorts and animal models, this translational study uncovers a microbial metabolite that might enhance treatment outcomes for notoriously difficult-to-treat pancreatic cancer.
Bender, M. J. et al. Dietary tryptophan metabolite released by intratumoral Lactobacillus reuteri facilitates immune checkpoint inhibitor treatment. Cell 186, 1846–1862.e26 (2023). This mechanistic preclinical study unveils how a conventional probiotic translocates to melanoma tumours and promotes antitumour immunity through production of the tryptophan catabolite indole-3-aldehyde.
Salminen, S. et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667 (2021).
Battaglia, T. W. et al. A pan-cancer analysis of the microbiome in metastatic cancer. Cell 187, 2324–2335.e19 (2024). This is a large exploration of human metastatic tumour biopsy-associated bacteria using metagenomics, genomics and transcriptomics and their association with immune infiltration and resistance to therapy.
Ghaddar, B. et al. Tumor microbiome links cellular programs and immunity in pancreatic cancer. Cancer Cell 40, 1240–1253.e5 (2022).
Riquelme, E. et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806.e12 (2019).
Nejman, D. et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 368, 973–980 (2020).
Perez Escriva, P., Correia Tavares Bernardino, C. & Letellier, E. De-coding the complex role of microbial metabolites in cancer. Cell Rep. 44, 115358 (2025).
El Tekle, G. & Garrett, W. S. Bacteria in cancer initiation, promotion and progression. Nat. Rev. Cancer 23, 600–618 (2023).
Narunsky-Haziza, L. et al. Pan-cancer analyses reveal cancer-type-specific fungal ecologies and bacteriome interactions. Cell 185, 3789–3806.e17 (2022).
Dohlman, A. B., Pan, X., Zitvogel, L. & Iliev, I. D. The multi-kingdom cancer microbiome. Nat. Microbiol. 10, 2369–2383 (2025).
Chagneau, C. V. et al. Uropathogenic E. coli induces DNA damage in the bladder. PLoS Pathog. 17, e1009310 (2021).
Dejea, C. M. et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359, 592–597 (2018). This study demonstrates that colonic biofilms of some patients with cancer often contain E. coli (producing colibactin) and enterotoxigenic. B. fragilis (producing BFT), whose genotoxins synergistically induce DNA damage and promote colorectal tumorigenesis in animal models.
Greathouse, K. L. et al. Interaction between the microbiome and TP53 in human lung cancer. Genome Biol. 19, 123 (2018).
Cullin, N., Azevedo Antunes, C., Straussman, R., Stein-Thoeringer, C. K. & Elinav, E. Microbiome and cancer. Cancer Cell 39, 1317–1341 (2021).
Shrestha, E. et al. Oncogenic gene fusions in nonneoplastic precursors as evidence that bacterial infection can initiate prostate cancer. Proc. Natl Acad. Sci. USA 118, e2018976118 (2021).
Maklin, T. et al. Geographical variation in the incidence of colorectal cancer and urinary tract cancer is associated with population exposure to colibactin-producing Escherichia coli. Lancet Microbe 6, 101015 (2025).
Kienesberger, S. et al. Enterotoxin tilimycin from gut-resident Klebsiella promotes mutational evolution and antibiotic resistance in mice. Nat. Microbiol. 7, 1834–1848 (2022).
Cao, Y. et al. Commensal microbiota from patients with inflammatory bowel disease produce genotoxic metabolites. Science 378, eabm3233 (2022).
Unterhauser, K. et al. Klebsiella oxytoca enterotoxins tilimycin and tilivalline have distinct host DNA-damaging and microtubule-stabilizing activities. Proc. Natl Acad. Sci. USA 116, 3774–3783 (2019).
Diaz-Gay, M. et al. Geographic and age variations in mutational processes in colorectal cancer. Nature https://doi.org/10.1038/s41586-025-09025-8 (2025). This study reports how mutational patterns in colorectal cancer vary by geographical and age variations, and it also highlights the role of colibactin in early-onset colorectal cancer.
Wilson, M. R. et al. The human gut bacterial genotoxin colibactin alkylates DNA. Science 363, eaar7785 (2019).
Oliero, M. et al. Prevalence of pks + bacteria and enterotoxigenic Bacteroides fragilis in patients with colorectal cancer. Gut Pathog. 14, 51 (2022).
Pleguezuelos-Manzano, C. et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020).
Rosendahl Huber, A. et al. Improved detection of colibactin-induced mutations by genotoxic E. coli in organoids and colorectal cancer. Cancer Cell 42, 487–496.e6 (2024).
Uemura, N. et al. Helicobacter pylori infection and the development of gastric cancer. N. Engl. J. Med. 345, 784–789 (2001).
Scanu, T. et al. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe 17, 763–774 (2015).
Niwa, T. et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 70, 1430–1440 (2010).
Wiklund, A. K. et al. Risk of gastric adenocarcinoma after eradication of Helicobacter pylori. Gastroenterology 169, 244–250.e1 (2025).
Jung, Y. S., Tran, M. T. X., Park, B. & Moon, C. M. Preventive effect of Helicobacter pylori treatment on gastric cancer incidence and mortality: a Korean population study. Gastroenterology 169, 251–260.e4 (2025).
Galeano Nino, J. L. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature 611, 810–817 (2022).
Zepeda-Rivera, M. et al. A distinct Fusobacterium nucleatum clade dominates the colorectal cancer niche. Nature 628, 424–432 (2024).
Zhou, R. W., Harpaz, N., Itzkowitz, S. H. & Parsons, R. E. Molecular mechanisms in colitis-associated colorectal cancer. Oncogenesis 12, 48 (2023).
Hajjar, R. et al. Basal levels of microbiota-driven subclinical inflammation are associated with anastomotic leak in patients with colorectal cancer. Gut 73, 1031–1033 (2024).
Hajjar, R. et al. Gut microbiota influence anastomotic healing in colorectal cancer surgery through modulation of mucosal proinflammatory cytokines. Gut 72, 1143–1154 (2023). This translational study shows how the gut microbiota modulates mucosal pro-inflammatory cytokines and, thereby, influences anastomotic healing after colorectal cancer surgery.
Hajjar, R. et al. Modulating gut microbiota prevents anastomotic leak to reduce local implantation and dissemination of colorectal cancer cells after surgery. Clin. Cancer Res. 30, 616–628 (2024).
Goodwin, A. C. et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. Proc. Natl Acad. Sci. USA 108, 15354–15359 (2011).
Fu, K. et al. Streptococcus anginosus promotes gastric inflammation, atrophy, and tumorigenesis in mice. Cell 187, 882–896.e17 (2024).
Inaba, Y. et al. Expression of the antimicrobial peptide α-defensin/cryptdins in intestinal crypts decreases at the initial phase of intestinal inflammation in a model of inflammatory bowel disease, IL-10-deficient mice. Inflamm. Bowel Dis. 16, 1488–1495 (2010).
Arthur, J. C. et al. Intestinal inflammation targets cancer-inducing activity of the microbiota. Science 338, 120–123 (2012).
Hajjar, R., Richard, C. & Santos, M. M. The gut barrier as a gatekeeper in colorectal cancer treatment. Oncotarget 15, 562–572 (2024).
Li, D. K. et al. Inhibition of microbial deconjugation of micellar bile acids protects against intestinal permeability and liver injury. Sci. Adv. 8, eabo2794 (2022).
Rahal, Z. et al. Inflammation mediated by gut microbiome alterations promotes lung cancer development and an immunosuppressed tumor microenvironment. Cancer Immunol. Res. 12, 1736–1752 (2024).
Zhang, Q. et al. Gut microbiome directs hepatocytes to recruit MDSCs and promote cholangiocarcinoma. Cancer Discov. 11, 1248–1267 (2021).
Hajjar, R., Richard, C. S. & Santos, M. M. The role of butyrate in surgical and oncological outcomes in colorectal cancer. Am. J. Physiol. Gastrointest. Liver Physiol. 320, G601–G608 (2021).
Yang, L. et al. Dietary folate and cofactors accelerate age-dependent p16 epimutation to promote intestinal tumorigenesis. Cancer Res. Commun. 4, 164–169 (2024).
Qin, Y. et al. An obesity-associated gut microbiome reprograms the intestinal epigenome and leads to altered colonic gene expression. Genome Biol. 19, 7 (2018).
Allen, J. et al. Colon tumors in enterotoxigenic Bacteroides fragilis (ETBF)-colonized mice do not display a unique mutational signature but instead possess host-dependent alterations in the APC gene. Microbiol. Spectr. 10, e0105522 (2022).
DeStefano Shields, C. E. et al. Bacterial-driven inflammation and mutant BRAF expression combine to promote murine colon tumorigenesis that is sensitive to immune checkpoint therapy. Cancer Discov. 11, 1792–1807 (2021).
Yang, Y. et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-κB, and up-regulating expression of microRNA-21. Gastroenterology 152, 851–866.e24 (2017).
Ansari, I. et al. The microbiota programs DNA methylation to control intestinal homeostasis and inflammation. Nat. Microbiol. 5, 610–619 (2020).
Korpela, K. et al. Selective maternal seeding and environment shape the human gut microbiome. Genome Res. 28, 561–568 (2018).
Nunez, H. et al. Early life gut microbiome and its impact on childhood health and chronic conditions. Gut Microbes 17, 2463567 (2025).
Reyman, M. et al. Effects of early-life antibiotics on the developing infant gut microbiome and resistome: a randomized trial. Nat. Commun. 13, 893 (2022).
Blaser, M. J. The theory of disappearing microbiota and the epidemics of chronic diseases. Nat. Rev. Immunol. 17, 461–463 (2017).
Lam, K. C. et al. Microbiota triggers STING-type I IFN-dependent monocyte reprogramming of the tumor microenvironment. Cell 184, 5338–5356.e21 (2021).
Vatanen, T. et al. The human gut microbiome in early-onset type 1 diabetes from the TEDDY study. Nature 562, 589–594 (2018).
Agrawal, M. et al. Early life exposures and the risk of inflammatory bowel disease: systematic review and meta-analyses. eClinicalMedicine 36, 100884 (2021).
Peppas, I., Ford, A. M., Furness, C. L. & Greaves, M. F. Gut microbiome immaturity and childhood acute lymphoblastic leukaemia. Nat. Rev. Cancer 23, 565–576 (2023).
Zhang, J. et al. Oral antibiotic use and risk of colorectal cancer in the United Kingdom, 1989-2012: a matched case-control study. Gut 68, 1971–1978 (2019).
Lu, S. S. M. et al. Antibiotics use and subsequent risk of colorectal cancer: a Swedish nationwide population-based study. J. Natl Cancer Inst. 114, 38–46 (2022).
Zimmermann, M., Zimmermann-Kogadeeva, M., Wegmann, R. & Goodman, A. L. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019).
Klunemann, M. et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 597, 533–538 (2021).
Spanogiannopoulos, P. et al. Host and gut bacteria share metabolic pathways for anti-cancer drug metabolism. Nat. Microbiol. 7, 1605–1620 (2022).
Liu, D. et al. Anaerostipes hadrus, a butyrate-producing bacterium capable of metabolizing 5-fluorouracil. mSphere 9, e0081623 (2024).
Javdan, B. et al. Personalized mapping of drug metabolism by the human gut microbiome. Cell 181, 1661–1679.e22 (2020).
Geller, L. T. et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 357, 1156–1160 (2017).
Trepka, K. R. et al. Expansion of a bacterial operon during cancer treatment ameliorates fluoropyrimidine toxicity. Sci. Transl. Med. 17, eadq8870 (2025).
Wallace, B. D. et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010).
LaCourse, K. D. et al. The cancer chemotherapeutic 5-fluorouracil is a potent Fusobacterium nucleatum inhibitor and its activity is modified by intratumoral microbiota. Cell Rep. 41, 111625 (2022).
McQuade, R. M., Al Thaalibi, M. & Nurgali, K. Impact of chemotherapy-induced enteric nervous system toxicity on gastrointestinal mucositis. Curr. Opin. Support. Palliat. Care 14, 293–300 (2020).
Xie, M., Li, X., Lau, H. C. & Yu, J. The gut microbiota in cancer immunity and immunotherapy. Cell Mol. Immunol. 22, 1012–1031 (2025).
Sharma, P. et al. Immune checkpoint therapy — current perspectives and future directions. Cell 186, 1652–1669 (2023).
Raskov, H., Orhan, A., Christensen, J. P. & Gogenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer 124, 359–367 (2021).
Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science 348, 56–61 (2015).
Matson, V. et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018).
Gopalakrishnan, V. et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018).
Derosa, L. et al. Intestinal Akkermansia muciniphila predicts clinical response to PD-1 blockade in patients with advanced non-small-cell lung cancer. Nat. Med. 28, 315–324 (2022). This study combines human and mouse evidence, demonstrating that intermediate (but not low or high) levels of A. muciniphila enhance PD1 blockade efficacy via promotion of T cell-mediated antitumour immunity.
Gunjur, A. et al. A gut microbial signature for combination immune checkpoint blockade across cancer types. Nat. Med. 30, 797–809 (2024).
Sivan, A. et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015).
Derosa, L. et al. Custom scoring based on ecological topology of gut microbiota associated with cancer immunotherapy outcome. Cell 187, 3373–3389.e16 (2024).
Mager, L. F. et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489 (2020).
Griffin, M. E. et al. Enterococcus peptidoglycan remodeling promotes checkpoint inhibitor cancer immunotherapy. Science 373, 1040–1046 (2021).
He, Y. et al. Gut microbial metabolites facilitate anticancer therapy efficacy by modulating cytotoxic CD8+ T cell immunity. Cell Metab. 33, 988–1000.e7 (2021).
Zhu, X. et al. Microbial metabolite butyrate promotes anti-PD-1 antitumor efficacy by modulating T cell receptor signaling of cytotoxic CD8 T cell. Gut Microbes 15, 2249143 (2023).
Luu, M. et al. Microbial short-chain fatty acids modulate CD8+ T cell responses and improve adoptive immunotherapy for cancer. Nat. Commun. 12, 4077 (2021).
He, L. et al. Gut microbiota-derived butyrate restores impaired regulatory T cells in patients with AChR myasthenia gravis via mTOR-mediated autophagy. Cell Commun. Signal. 22, 215 (2024).
Coutzac, C. et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 11, 2168 (2020).
Xiong, Z. et al. Targeting PPAR-gamma counteracts tumour adaptation to immune-checkpoint blockade in hepatocellular carcinoma. Gut 72, 1758–1773 (2023).
Damotte, D. et al. The tumor inflammation signature (TIS) is associated with anti-PD-1 treatment benefit in the CERTIM pan-cancer cohort. J. Transl. Med. 17, 357 (2019).
Sui, Q. et al. Inflammation promotes resistance to immune checkpoint inhibitors in high microsatellite instability colorectal cancer. Nat. Commun. 13, 7316 (2022).
Huang, J. et al. Ginseng polysaccharides alter the gut microbiota and kynurenine/tryptophan ratio, potentiating the antitumour effect of antiprogrammed cell death 1/programmed cell death ligand 1 (anti-PD-1/PD-L1) immunotherapy. Gut 71, 734–745 (2022).
Qin, R. et al. Tryptophan potentiates CD8+ T cells against cancer cells by TRIP12 tryptophanylation and surface PD-1 downregulation. J. Immunother. Cancer 9, e002840 (2021).
Jia, D. et al. Microbial metabolite enhances immunotherapy efficacy by modulating T cell stemness in pan-cancer. Cell 187, 1651–1665.e21 (2024). This preclinical study demonstrates that the gut microbial metabolite indole-3-propionic acid broadly enhanced the effectiveness of immune checkpoint blockade across diverse cancer types by epigenetically programming CD8+ T cells into a stem-like, therapeutically responsive state.
Alves Costa Silva, C. et al. Influence of microbiota-associated metabolic reprogramming on clinical outcome in patients with melanoma from the randomized adjuvant dendritic cell-based MIND-DC trial. Nat. Commun. 15, 1633 (2024).
Uribe-Herranz, M. et al. Gut microbiota modulates adoptive cell therapy via CD8α dendritic cells and IL-12. JCI Insight 3, e94952 (2018).
Smith, M. et al. Gut microbiome correlates of response and toxicity following anti-CD19 CAR T cell therapy. Nat. Med. 28, 713–723 (2022).
Marcos-Kovandzic, L. et al. Gut microbiota modulation through Akkermansia spp. supplementation increases CAR T-cell potency. Cancer Discov. 15, 1905–1926 (2025).
Khuat, L. T., Dave, M. & Murphy, W. J. The emerging roles of the gut microbiome in allogeneic hematopoietic stem cell transplantation. Gut Microbes 13, 1966262 (2021).
Chen, J., Deutsch, E., Kroemer, G., Galluzzi, L. & Zitvogel, L. The microbiota in radiotherapy-induced cancer immunosurveillance. Nat. Rev. Clin. Oncol. 22, 667–679 (2025).
Ji, M. et al. Methionine restriction-induced sulfur deficiency impairs antitumour immunity partially through gut microbiota. Nat. Metab. 5, 1526–1543 (2023).
Oh, M. H. et al. Targeting glutamine metabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J. Clin. Invest. 130, 3865–3884 (2020).
Cuisiniere, T. et al. Initial gut microbiota composition is a determining factor in the promotion of colorectal cancer by oral iron supplementation: evidence from a murine model. Microbiome 13, 100 (2025).
Westdorp, H. et al. Mechanisms of immune checkpoint inhibitor-mediated colitis. Front. Immunol. 12, 768957 (2021).
Gao, Y. Q., Tan, Y. J. & Fang, J. Y. Roles of the gut microbiota in immune-related adverse events: mechanisms and therapeutic intervention. Nat. Rev. Clin. Oncol. 22, 499–516 (2025).
Andrews, M. C. et al. Gut microbiota signatures are associated with toxicity to combined CTLA-4 and PD-1 blockade. Nat. Med. 27, 1432–1441 (2021).
Hu, M. et al. Gut microbiome for predicting immune checkpoint blockade-associated adverse events. Genome Med. 16, 16 (2024).
Liu, W. et al. Intestinal microbiome associated with immune-related adverse events for patients treated with anti-PD-1 inhibitors, a real-world study. Front. Immunol. 12, 756872 (2021).
Dubin, K. et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat. Commun. 7, 10391 (2016).
Cascone, T. et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: the phase 2 randomized NEOSTAR trial. Nat. Med. 27, 504–514 (2021).
McCulloch, J. A. et al. Intestinal microbiota signatures of clinical response and immune-related adverse events in melanoma patients treated with anti-PD-1. Nat. Med. 28, 545–556 (2022).
Bjork, J. R. et al. Longitudinal gut microbiome changes in immune checkpoint blockade-treated advanced melanoma. Nat. Med. 30, 785–796 (2024).
Lo, J. W. et al. Immune checkpoint inhibitor-induced colitis is mediated by polyfunctional lymphocytes and is dependent on an IL23/IFNγ axis. Nat. Commun. 14, 6719 (2023).
Lo, B. C. et al. Microbiota-dependent activation of CD4+ T cells induces CTLA-4 blockade-associated colitis via Fcγ receptors. Science 383, 62–70 (2024). This study introduces a preclinical model for ICI-colitis by colonizing mice with wild rodent gut microbiota, which enabled robust development of CTLA4 blockade-associated colitis.
Gao, Y. et al. Faecalibacterium prausnitzii abrogates intestinal toxicity and promotes tumor immunity to increase the efficacy of dual CTLA4 and PD-1 checkpoint blockade. Cancer Res. 83, 3710–3725 (2023).
Wang, T. et al. Probiotics Lactobacillus reuteri abrogates immune checkpoint blockade-associated colitis by inhibiting group 3 innate lymphoid cells. Front. Immunol. 10, 1235 (2019).
Yi, Y. et al. Gut microbiome components predict response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer: a prospective, longitudinal study. Clin. Cancer Res. 27, 1329–1340 (2021).
Shaikh, F. Y. et al. Fecal microbiome composition correlates with pathologic complete response in patients with operable esophageal cancer treated with combined chemoradiotherapy and immunotherapy. Cancers 16, 3644 (2024).
Viaud, S. et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013).
Vatanen, T. et al. Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 165, 842–853 (2016).
Sulit, A. K. et al. Bacterial lipopolysaccharide modulates immune response in the colorectal tumor microenvironment. NPJ Biofilms Microbiomes 9, 59 (2023).
Hu, Y. et al. CAR-T cell therapy-related cytokine release syndrome and therapeutic response is modulated by the gut microbiome in hematologic malignancies. Nat. Commun. 13, 5313 (2022).
Fertitta, V. et al. Akkermansia muciniphila- and pathogenic bacteria-derived endotoxins differently regulate human dendritic cell generation and γδ T lymphocyte activation. Biomolecules 14, 1571 (2024).
Roberti, M. P. et al. Chemotherapy-induced ileal crypt apoptosis and the ileal microbiome shape immunosurveillance and prognosis of proximal colon cancer. Nat. Med. 26, 919–931 (2020).
Louie, T. et al. VE303, a defined bacterial consortium, for prevention of recurrent Clostridioides difficile infection: a randomized clinical trial. JAMA 329, 1356–1366 (2023).
Tanoue, T. et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature 565, 600–605 (2019).
Kurt, F. et al. Co-cultivation is a powerful approach to produce a robust functionally designed synthetic consortium as a live biotherapeutic product (LBP). Gut Microbes 15, 2177486 (2023).
Menon, R. et al. Multi-omic profiling a defined bacterial consortium for treatment of recurrent Clostridioides difficile infection. Nat. Med. 31, 223–234 (2025).
Kang, Z. et al. A polyvalent vaccine for selectively killing tumor-associated bacteria to prevent cancer metastasis. Sci. Adv. 11, eadt0341 (2025).
Diamantis, S. et al. The production of antibiotics must be reoriented: repositioning old narrow-spectrum antibiotics, developing new microbiome-sparing antibiotics. Antibiotics 11, 924 (2022).
Ritz, N. L. et al. The gut virome is associated with stress-induced changes in behaviour and immune responses in mice. Nat. Microbiol. 9, 359–376 (2024).
Zechner, E. L. & Kienesberger, S. Microbiota-derived small molecule genotoxins: host interactions and ecological impact in the gut ecosystem. Gut Microbes 16, 2430423 (2024).
Bossuet-Greif, N. et al. Escherichia coli ClbS is a colibactin resistance protein. Mol. Microbiol. 99, 897–908 (2016).
Tripathi, P. et al. ClbS is a cyclopropane hydrolase that confers colibactin resistance. J. Am. Chem. Soc. 139, 17719–17722 (2017).
Oliero, M. et al. Putrescine supplementation limits the expansion of pks+ Escherichia coli and tumor development in the colon. Cancer Res. Commun. 4, 1777–1792 (2024).
Ledala, N. et al. Bacterial indole as a multifunctional regulator of Klebsiella oxytoca complex enterotoxicity. mBio 13, e0375221 (2022).
Volpe, M. R. et al. A small molecule inhibitor prevents gut bacterial genotoxin production. Nat. Chem. Biol. 19, 159–167 (2023).
Lopez-Romero, D. et al. Evidence of some natural products with antigenotoxic effects. Part 2: plants, vegetables, and natural resin. Nutrients 10, 1954 (2018).
Chou, P. J. et al. Epigenetics of dietary phytochemicals in cancer prevention: fact or fiction. Cancer J. 30, 320–328 (2024).
Yin, Q. et al. Ecological dynamics of Enterobacteriaceae in the human gut microbiome across global populations. Nat. Microbiol. 10, 541–553 (2025).
Hecht, A. L. et al. Dietary carbohydrates regulate intestinal colonization and dissemination of Klebsiella pneumoniae. J. Clin. Invest. 134, e174726 (2024).
Thakur, B. K. et al. Dietary fibre counters the oncogenic potential of colibactin-producing Escherichia coli in colorectal cancer. Nat. Microbiol. 10, 855–870 (2025).
Furuichi, M. et al. Commensal consortia decolonize Enterobacteriaceae via ecological control. Nature 633, 878–886 (2024).
Jacobs, J. P. et al. Human milk oligosaccharides modulate the intestinal microbiome of healthy adults. Sci. Rep. 13, 14308 (2023).
Ervin, S. M. et al. Targeting regorafenib-induced toxicity through inhibition of gut microbial β-glucuronidases. ACS Chem. Biol. 14, 2737–2744 (2019).
Chamseddine, A. N. et al. Intestinal bacterial β-glucuronidase as a possible predictive biomarker of irinotecan-induced diarrhea severity. Pharmacol. Ther. 199, 1–15 (2019).
Ianiro, G. et al. Faecal microbiota transplantation for the treatment of diarrhoea induced by tyrosine-kinase inhibitors in patients with metastatic renal cell carcinoma. Nat. Commun. 11, 4333 (2020).
Mego, M. et al. Prevention of irinotecan induced diarrhea by probiotics: a randomized double blind, placebo controlled pilot study. Complement. Ther. Med. 23, 356–362 (2015).
Davar, D. et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 371, 595–602 (2021). This phase I clinical trial demonstrates that FMT from anti-PD1 responders might overcome resistance to anti-PD1 therapy in patients with melanoma who had not responded to ICIs previously.
Baruch, E. N. et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 371, 602–609 (2021).
Routy, B. et al. Fecal microbiota transplantation plus anti-PD-1 immunotherapy in advanced melanoma: a phase I trial. Nat. Med. 29, 2121–2132 (2023).
Ciccarese, C. et al. LBA77 fecal microbiota transplantation (FMT) versus placebo in patients receiving pembrolizumab plus axitinib for metastatic renal cell carcinoma: preliminary results of the randomized phase II TACITO trial. Ann. Oncol. 35, 51264 (2024).
Wang, Y. et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat. Med. 24, 1804–1808 (2018). This pilot study shows that FMT can be efficient in alleviating ICI-associated colitis.
Duttagupta, S. et al. Microbiome profiling reveals that fecal microbiota transplantation (FMT) modulates response and toxicity when combined with immunotherapy in patients with lung cancer and melanoma (FMT-LUMINate NCT04951583). Cancer Res. 85, abstr. 2210 (2025).
Spencer, C. N. et al. Dietary fiber and probiotics influence the gut microbiome and melanoma immunotherapy response. Science 374, 1632–1640 (2021).
Zhang, X. et al. Modulating a prebiotic food source influences inflammation and immune-regulating gut microbes and metabolites: insights from the BE GONE trial. eBioMedicine 98, 104873 (2023).
Farias, R. M. et al. Diet and Immune Effects Trial (DIET) — a randomized, double-blinded dietary intervention study in patients with melanoma receiving immunotherapy. BMC Cancer 24, 1493 (2024).
Jiang, Y. et al. The DIET study: a randomized controlled trial of a high fiber diet intervention (HFDI) in patients (pts) with melanoma receiving immune checkpoint blockade (ICB). J. Clin. Oncol. https://doi.org/10.1200/JCO.2024.42.16_suppl.9558 (2024).
Ferrere, G. et al. Ketogenic diet and ketone bodies enhance the anticancer effects of PD-1 blockade. JCI Insight 6, e145207 (2021).
Plamada, D. & Vodnar, D. C. Polyphenols–gut microbiota interrelationship: a transition to a new generation of prebiotics. Nutrients 14, 137 (2021).
Messaoudene, M. et al. A natural polyphenol exerts antitumor activity and circumvents anti-PD-1 resistance through effects on the gut microbiota. Cancer Discov. 12, 1070–1087 (2022).
Giampazolias, E. et al. Vitamin D regulates microbiome-dependent cancer immunity. Science 384, 428–437 (2024).
Qureshi, Z., Jamil, A., Altaf, F. & Siddique, R. Efficacy and safety of probiotics as adjunctive therapy in cancer treatment: a comprehensive systematic review and meta-analysis. Am. J. Clin. Oncol. 48, 148–161 (2025).
Liu, Y. C., Wu, C. R. & Huang, T. W. Preventive effect of probiotics on oral mucositis induced by cancer treatment: a systematic review and meta-analysis. Int. J. Mol. Sci. 23, 13268 (2022).
Han, Y., Wang, Y. & Guan, M. Preventive effect of probiotics on infections following colorectal cancer surgery: an umbrella meta-analysis. World J. Gastrointest. Surg. 16, 3546–3558 (2024).
Zhao, S. et al. Assessing the impact of probiotics on immunotherapy effectiveness and antibiotic-mediated resistance in cancer: a systematic review and meta-analysis. Front. Immunol. 16, 1538969 (2025).
Fong, W. et al. Lactobacillus gallinarum-derived metabolites boost anti-PD1 efficacy in colorectal cancer by inhibiting regulatory T cells through modulating IDO1/Kyn/AHR axis. Gut 72, 2272–2285 (2023).
Redenti, A. et al. Probiotic neoantigen delivery vectors for precision cancer immunotherapy. Nature 635, 453–461 (2024).
Kwon, S. Y., Thi-Thu Ngo, H., Son, J., Hong, Y. & Min, J. J. Exploiting bacteria for cancer immunotherapy. Nat. Rev. Clin. Oncol. 21, 569–589 (2024).
Canale, F. P. et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature 598, 662–666 (2021). This preclinical study reports that engineered probiotic bacteria that convert intratumoural ammonia into l-arginine can locally boost cytotoxic T cell immunity and amplify checkpoint therapy effects.
Geiger, R. et al. L-Arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842.e13 (2016).
Zhao, H. et al. Inosine enhances the efficacy of immune-checkpoint inhibitors in advanced solid tumors: a randomized, controlled, phase 2 study. Cancer Med. 13, e70143 (2024). Building on an earlier preclinical work on gut-derived inosine, this rare phase II randomized clinical trial has found that oral supplementation of inosine significantly enhanced ICI efficacy in patients with solid tumours.
Talpin, A. et al. Mimicry-based strategy between human and commensal antigens for the development of a new family of immune therapies for cancer. J. Immunother. Cancer 13, e010192 (2025).
Courneya, K. S. et al. Structured exercise after adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 393, 13–25 (2025).
Phelps, C. M. et al. Exercise-induced microbiota metabolite enhances CD8 T cell antitumor immunity promoting immunotherapy efficacy. Cell https://doi.org/10.1016/j.cell.2025.06.018 (2025).
Thomas, A. M. et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 25, 667–678 (2019).
Wirbel, J. et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 25, 679–689 (2019).
Tito, R. Y. et al. Microbiome confounders and quantitative profiling challenge predicted microbial targets in colorectal cancer development. Nat. Med. 30, 1339–1348 (2024).
Fierer, N. et al. Guidelines for preventing and reporting contamination in low-biomass microbiome studies. Nat. Microbiol. 10, 1570–1580 (2025).
Gihawi, A. et al. Major data analysis errors invalidate cancer microbiome findings. mBio 14, e0160723 (2023).
Austin, G. I. & Korem, T. Compositional transformations can reasonably introduce phenotype-associated values into sparse features. mSystems 10, e0002125 (2025).
Sepich-Poore, G. D. et al. Robustness of cancer microbiome signals over a broad range of methodological variation. Oncogene 43, 1127–1148 (2024).
Ge, Y., Lu, J., Puiu, D., Revsine, M. & Salzberg, S. L. Comprehensive analysis of microbial content in whole-genome sequencing samples from The Cancer Genome Atlas project. Sci. Transl. Med. 17, eads6335 (2025).
Chang, D. et al. Gut Microbiome Wellness Index 2 enhances health status prediction from gut microbiome taxonomic profiles. Nat. Commun. 15, 7447 (2024).
Wu, G. et al. A core microbiome signature as an indicator of health. Cell 187, 6550–6565.e11 (2024).
Yonekura, S. et al. Cancer induces a stress ileopathy depending on β-adrenergic receptors and promoting dysbiosis that contributes to carcinogenesis. Cancer Discov. 12, 1128–1151 (2022). This translational study proposes that many cancers could have a shared effect on the gut microbiome, which might then act as a modulator of subsequent disease course.
Kartal, E. et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut 71, 1359–1372 (2022). This research shows an example of a high-quality faecal microbiota classifier that has been developed to detect pancreatic cancer and has been validated across multiple independent cohorts and external datasets.
Simpson, R. C. et al. Diet-driven microbial ecology underpins associations between cancer immunotherapy outcomes and the gut microbiome. Nat. Med. 28, 2344–2352 (2022).
Glitza, I. C. et al. Randomized placebo-controlled, biomarker-stratified phase Ib microbiome modulation in melanoma: impact of antibiotic preconditioning on microbiome and immunity. Cancer Discov. 14, 1161–1175 (2024).
Yin, Q. et al. Immune-related adverse events of immune checkpoint inhibitors: a review. Front. Immunol. 14, 1167975 (2023).
Wu, G., Zhao, N., Zhang, C., Lam, Y. Y. & Zhao, L. Guild-based analysis for understanding gut microbiome in human health and diseases. Genome Med. 13, 22 (2021).
Muller, E., Shiryan, I. & Borenstein, E. Multi-omic integration of microbiome data for identifying disease-associated modules. Nat. Commun. 15, 2621 (2024).
Wang, N. et al. MSFT-transformer: a multistage fusion tabular transformer for disease prediction using metagenomic data. Brief. Bioinform. 26, bbaf217 (2025).
Hall, A. B. et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med. 9, 103 (2017).
Hernandez Medina, R. et al. Machine learning and deep learning applications in microbiome research. ISME Commun. 2, 98 (2022).
Dang, T., Lysenko, A., Boroevich, K. A. & Tsunoda, T. VBayesMM: variational Bayesian neural network to prioritize important relationships of high-dimensional microbiome multiomics data. Brief. Bioinform. 26, bbaf300 (2025).
Ding, Y. et al. Identification of gut microbial bile acid metabolic enzymes via an AI-assisted pipeline. Cell https://doi.org/10.1016/j.cell.2025.07.017 (2025).
Tang, X. et al. OmicsFootPrint: a framework to integrate and interpret multi-omics data using circular images and deep neural networks. Nucleic Acids Res. 52, e99 (2024).
Mock, F., Kretschmer, F., Kriese, A., Bocker, S. & Marz, M. Taxonomic classification of DNA sequences beyond sequence similarity using deep neural networks. Proc. Natl Acad. Sci. USA 119, e2122636119 (2022).
Consens, M. E. et al. Transformers and genome language models. Nat. Mach. Intell. 7, 346–362 (2025).
De Domenico, M. et al. Challenges and opportunities for digital twins in precision medicine from a complex systems perspective. NPJ Digit. Med. 8, 37 (2025).
Sizemore, N. et al. A digital twin of the infant microbiome to predict neurodevelopmental deficits. Sci. Adv. 10, eadj0400 (2024).
Tripathi, P. & Bruner, S. D. Structural basis for the interactions of the colibactin resistance gene product ClbS with DNA. Biochemistry 60, 1619–1625 (2021).
Chagneau, C. V. et al. The polyamine spermidine modulates the production of the bacterial genotoxin colibactin. mSphere 4, e00414–e00419 (2019).
Minot, S. S. et al. Colorectal cancer-associated bacteria are broadly distributed in global microbiomes and drivers of precancerous change. Sci. Rep. 14, 23646 (2024).
Roje, B. et al. Gut microbiota carcinogen metabolism causes distal tissue tumours. Nature 632, 1137–1144 (2024). This preclinical study reveals that gut bacteria metabolize nitrosamine carcinogens into oxidized metabolites that accumulate in distal organs beyond the gastrointestinal tract, thereby increasing tumour susceptibility in these tissues.
Ervin, S. M. et al. Gut microbial β-glucuronidases reactivate estrogens as components of the estrobolome that reactivate estrogens. J. Biol. Chem. 294, 18586–18599 (2019).
Hebels, D. G. et al. N-Nitroso compound exposure-associated transcriptomic profiles are indicative of an increased risk for colorectal cancer. Cancer Lett. 309, 1–10 (2011).
Meyer, C. et al. Chronic inflammation promotes myeloid-derived suppressor cell activation blocking antitumor immunity in transgenic mouse melanoma model. Proc. Natl Acad. Sci. USA 108, 17111–17116 (2011).
Ternes, D. et al. The gut microbial metabolite formate exacerbates colorectal cancer progression. Nat. Metab. 4, 458–475 (2022).
Yu, T. et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell 170, 548–563.e16 (2017).
Gao, Y. et al. Fusobacterium nucleatum enhances the efficacy of PD-L1 blockade in colorectal cancer. Signal Transduct. Target. Ther. 6, 398 (2021).
Abdeen, S. K., Mastandrea, I., Stinchcombe, N., Puschhof, J. & Elinav, E. Diet-microbiome interactions in cancer. Cancer Cell 43, 680–707 (2025).
Bagheri, A., Asoudeh, F., Rezaei, S., Babaei, M. & Esmaillzadeh, A. The effect of Mediterranean diet on body composition, inflammatory factors, and nutritional status in patients with cachexia induced by colorectal cancer: a randomized clinical trial. Integr. Cancer Ther. 22, 15347354231195322 (2023).
Acknowledgements
The authors thank L. Busby (Mayo Clinic, Rochester, MN, USA) for the administrative assistance.
Author information
Authors and Affiliations
Contributions
All authors researched data for the article, contributed substantially to the discussion of the content, wrote the article, and reviewed and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
P.C.K. is a member of the scientific advisory board of the International Observatory of Biocodex Microbiota Institute, chair of the scientific advisory board of the American Gastroenterology Association Center for Gut Microbiome Education and Research, ad hoc advisory board member for Pendulum and Intrinsic Medicine, and advisory board member for 32 Biosciences for which he receives equity option as compensation. R.A.T.M. serves as a founding adviser of Tiny Health. R.H. declares no competing interests.
Peer review
Peer review information
Nature Reviews Microbiology thanks Cynthia Sears and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hajjar, R., Mars, R.A.T. & Kashyap, P.C. Harnessing the microbiome for cancer therapy. Nat Rev Microbiol (2026). https://doi.org/10.1038/s41579-025-01268-6
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41579-025-01268-6