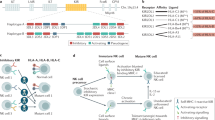
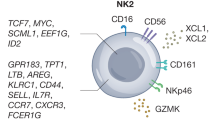
Abstract
Natural killer (NK) cells are cytotoxic lymphocytes of the innate immune system with essential roles in immune surveillance, tissue homeostasis and inflammation. In the kidney, NK cells comprise a heterogeneous population that includes both circulating and tissue-resident subsets, each shaped by environmental cues and genetic factors through interactions with major histocompatibility complex class I molecules. The latest research data highlight multifaceted NK cell contributions to kidney physiology and pathology. In steady state, NK cells support kidney immune surveillance and crosstalk with epithelial, myeloid and lymphoid cells. In disease, NK cells can promote injury through direct cytotoxicity and pro-inflammatory cytokine release. Experimental models demonstrate pathogenic roles for NK cells in ischaemia–reperfusion injury and chronic kidney disease, with emerging evidence implicating NK cell-derived mediators in fibrogenesis. In kidney transplantation, NK cells are effectors of antibody-dependent and antibody-independent allograft injury. Educated NK cells expressing CD16a (also known as FcγRIIIa) mediate antibody-dependent cellular cytotoxicity, whereas loss of inhibitory receptor–ligand interactions (for example, due to killer immunoglobulin-like receptor–HLA mismatch) can trigger NK cell activation independently of donor-specific antibodies. Advances in high-resolution profiling have deepened mechanistic insights and uncovered novel therapeutic targets. Here, we provide a comprehensive overview of NK cell biology in the kidney, highlighting roles in health, disease and transplantation, and we consider its translational implications for diagnosis and therapy.
Key points
-
Natural killer (NK) cells are integral to kidney immune surveillance, maintaining homeostasis through cytokine production, interactions with epithelial cells and crosstalk with other immune cells, including macrophages and dendritic cells.
-
Tissue-resident NK cells adapt to the kidney microenvironment, acquiring unique phenotypes and non-cytolytic functions, such as the expression of integrins and chemokines, that distinguish them from circulating NK cell subsets.
-
In acute kidney injury, NK cells initiate inflammation and promote epithelial damage via direct cytotoxicity and recruitment of pro-inflammatory myeloid cells.
-
In chronic kidney disease, NK cell-derived IFNγ, granzymes and 37 kDa killer-specific secretory protein can contribute to fibrosis through fibroblast activation and maladaptive tissue remodelling.
-
In transplantation, NK cells mediate both antibody-dependent and antibody-independent allograft injury, shaped by NK cell education, host genetics and history of viral exposure.
-
Targeting NK cells and their effector pathways (for example, by targeting IL-15, mechanistic target of rapamycin, CD38 and NKG2D) represents a promising therapeutic strategy to improve transplantation outcomes and ameliorate other forms of kidney disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lanier, L. L. Five decades of natural killer cell discovery. J. Exp. Med. 221, e20231222 (2024).
Vivier, E. et al. Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49 (2011).
Cerwenka, A. & Lanier, L. L. Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol. 16, 112–123 (2016).
Carrega, P. et al. CD56brightperforinlow noncytotoxic human NK cells are abundant in both healthy and neoplastic solid tissues and recirculate to secondary lymphoid organs via afferent lymph. J. Immunol. 192, 3805–3815 (2014).
Raulet, D. H., Marcus, A. & Coscoy, L. Dysregulated cellular functions and cell stress pathways provide critical cues for activating and targeting natural killer cells to transformed and infected cells. Immunol. Rev. 280, 93–101 (2017).
Horowitz, A. et al. Class I HLA haplotypes form two schools that educate NK cells in different ways. Sci. Immunol. 1, eaag1672 (2016).
Strauss-Albee, D. M., Horowitz, A., Parham, P. & Blish, C. A. Coordinated regulation of NK receptor expression in the maturing human immune system. J. Immunol. 193, 4871–4879 (2014).
Horowitz, A. et al. Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci. Transl. Med. 5, 208ra145 (2013).
Hegewisch-Solloa, E. et al. Mapping human natural killer cell development in pediatric tonsil by imaging mass cytometry and high-resolution microscopy. Preprint at bioRxiv https://doi.org/10.1101/2023.09.05.556371 (2023).
Freud, A. G. & Caligiuri, M. A. Human natural killer cell development. Immunol. Rev. 214, 56–72 (2006).
Freud, A. G. et al. Evidence for discrete stages of human natural killer cell differentiation in vivo. J. Exp. Med. 203, 1033–1043 (2006).
Bjorkstrom, N. K., Ljunggren, H. G. & Michaelsson, J. Emerging insights into natural killer cells in human peripheral tissues. Nat. Rev. Immunol. 16, 310–320 (2016).
Netskar, H. et al. Pan-cancer profiling of tumor-infiltrating natural killer cells through transcriptional reference mapping. Nat. Immunol. 25, 1445–1459 (2024).
Rebuffet, L. et al. High-dimensional single-cell analysis of human natural killer cell heterogeneity. Nat. Immunol. 25, 1474–1488 (2024).
Anfossi, N. et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity 25, 331–342 (2006).
Yokoyama, W. M. & Kim, S. Licensing of natural killer cells by self-major histocompatibility complex class I. Immunol. Rev. 214, 143–154 (2006).
Hoglund, P. et al. Recognition of beta 2-microglobulin-negative (beta 2m-) T-cell blasts by natural killer cells from normal but not from beta 2m- mice: nonresponsiveness controlled by beta 2m- bone marrow in chimeric mice. Proc. Natl Acad. Sci. USA 88, 10332–10336 (1991).
Brodin, P., Lakshmikanth, T., Johansson, S., Karre, K. & Hoglund, P. The strength of inhibitory input during education quantitatively tunes the functional responsiveness of individual natural killer cells. Blood 113, 2434–2441 (2009).
Held, W. & Raulet, D. H. Ly49A transgenic mice provide evidence for a major histocompatibility complex-dependent education process in natural killer cell development. J. Exp. Med. 185, 2079–2088 (1997).
Parham, P. & Guethlein, L. A. Genetics of natural killer cells in human health, disease, and survival. Annu. Rev. Immunol. 36, 519–548 (2018).
Parham, P. & Moffett, A. Variable NK cell receptors and their MHC class I ligands in immunity, reproduction and human evolution. Nat. Rev. Immunol. 13, 133–144 (2013).
Parham, P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 5, 201–214 (2005).
Kim, S. et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature 436, 709–713 (2005).
Migdal, M., Ruan, D. F., Forrest, W. F., Horowitz, A. & Hammer, C. MiDAS-meaningful immunogenetic data at scale. PLoS Comput. Biol. 17, e1009131 (2021).
Petersdorf, E. W. et al. HLA-B leader and survivorship after HLA-mismatched unrelated donor transplantation. Blood 136, 362–369 (2020).
Cooley, S., Parham, P. & Miller, J. S. Strategies to activate NK cells to prevent relapse and induce remission following hematopoietic stem cell transplantation. Blood 131, 1053–1062 (2018).
Martin, M. P. et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat. Genet. 39, 733–740 (2007).
Alter, G. et al. HIV-1 adaptation to NK-cell-mediated immune pressure. Nature 476, 96–100 (2011).
Khakoo, S. I. et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305, 872–874 (2004).
Ramsuran, V. et al. Elevated HLA-A expression impairs HIV control through inhibition of NKG2A-expressing cells. Science 359, 86–90 (2018).
Smith, S. L. et al. Diversity of peripheral blood human NK cells identified by single-cell RNA sequencing. Blood Adv. 4, 1388–1406 (2020).
Moretta, L. Dissecting CD56dim human NK cells. Blood 116, 3689–3691 (2010).
Michel, T. et al. Human CD56bright NK cells: an update. J. Immunol. 196, 2923–2931 (2016).
Poli, A. et al. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology 126, 458–465 (2009).
Nielsen, C. M., White, M. J., Goodier, M. R. & Riley, E. M. Functional significance of CD57 expression on human NK cells and relevance to disease. Front. Immunol. 4, 422 (2013).
Kared, H., Martelli, S., Ng, T. P., Pender, S. L. & Larbi, A. CD57 in human natural killer cells and T-lymphocytes. Cancer Immunol. Immunother. 65, 441–452 (2016).
Crinier, A., Narni-Mancinelli, E., Ugolini, S. & Vivier, E. SnapShot: natural killer cells. Cell 180, 1280–1280.e1 (2020).
Carlyle, J. R. et al. Molecular and genetic basis for strain-dependent NK1.1 alloreactivity of mouse NK cells. J. Immunol. 176, 7511–7524 (2006).
Melsen, J. E., Lugthart, G., Lankester, A. C. & Schilham, M. W. Human circulating and tissue-resident CD56bright natural killer cell populations. Front. Immunol. 7, 262 (2016).
Sun, K., Li, Y. Y. & Jin, J. A double-edged sword of immuno-microenvironment in cardiac homeostasis and injury repair. Signal Transduct. Target. Ther. 6, 79 (2021).
Krizhanovsky, V. et al. Senescence of activated stellate cells limits liver fibrosis. Cell 134, 657–667 (2008).
Fasbender, F., Widera, A., Hengstler, J. G. & Watzl, C. Natural killer cells and liver fibrosis. Front. Immunol. 7, 19 (2016).
Zaiatz-Bittencourt, V., Finlay, D. K. & Gardiner, C. M. Canonical TGF-β signaling pathway represses human NK cell metabolism. J. Immunol. 200, 3934–3941 (2018).
Assmann, N. et al. Srebp-controlled glucose metabolism is essential for NK cell functional responses. Nat. Immunol. 18, 1197–1206 (2017).
Loftus, R. M. et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat. Commun. 9, 2341 (2018).
Lozada, J. R., Zhang, B., Miller, J. S. & Cichocki, F. NK cells from human cytomegalovirus-seropositive individuals have a distinct metabolic profile that correlates with elevated mTOR signaling. J. Immunol. 211, 539–550 (2023).
Tang, F. et al. A pan-cancer single-cell panorama of human natural killer cells. Cell 186, 4235–4251.e20 (2023).
Sohn, H. & Cooper, M. A. Metabolic regulation of NK cell function: implications for immunotherapy. Immunometabolism 5, e00020 (2023).
Moretta, A. Natural killer cells and dendritic cells: rendezvous in abused tissues. Nat. Rev. Immunol. 2, 957–964 (2002).
Jabri, B. & Abadie, V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol. 15, 771–783 (2015).
Newman, K. C. & Riley, E. M. Whatever turns you on: accessory-cell-dependent activation of NK cells by pathogens. Nat. Rev. Immunol. 7, 279–291 (2007).
Cooper, M. A. et al. Human natural killer cells: a unique innate immunoregulatory role for the CD56bright subset. Blood 97, 3146–3151 (2001).
Cooper, M. A., Fehniger, T. A. & Caligiuri, M. A. The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640 (2001).
Kennedy, M. K. et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J. Exp. Med. 191, 771–780 (2000).
Waldmann, T. A. & Tagaya, Y. The multifaceted regulation of interleukin-15 expression and the role of this cytokine in NK cell differentiation and host response to intracellular pathogens. Annu. Rev. Immunol. 17, 19–49 (1999).
Huntington, N. D., Vosshenrich, C. A. & Di Santo, J. P. Developmental pathways that generate natural-killer-cell diversity in mice and humans. Nat. Rev. Immunol. 7, 703–714 (2007).
Ma, S., Caligiuri, M. A. & Yu, J. Harnessing IL-15 signaling to potentiate NK cell-mediated cancer immunotherapy. Trends Immunol. 43, 833–847 (2022).
Ataya, M. et al. Long-term evolution of the adaptive NKG2C+ NK cell response to cytomegalovirus infection in kidney transplantation: an insight on the diversity of host–pathogen interaction. J. Immunol. 207, 1882–1890 (2021).
Lopez-Botet, M. et al. Development of the adaptive NK cell response to human cytomegalovirus in the context of aging. Mech. Ageing Dev. 158, 23–26 (2016).
Guma, M. et al. Expansion of CD94/NKG2C+ NK cells in response to human cytomegalovirus-infected fibroblasts. Blood 107, 3624–3631 (2006).
Guma, M., Angulo, A. & Lopez-Botet, M. NK cell receptors involved in the response to human cytomegalovirus infection. Curr. Top. Microbiol. Immunol. 298, 207–223 (2006).
Beziat, V. et al. NK cell responses to cytomegalovirus infection lead to stable imprints in the human KIR repertoire and involve activating KIRs. Blood 121, 2678–2688 (2013).
Djaoud, Z. et al. Cytomegalovirus-infected primary endothelial cells trigger NKG2C+ natural killer cells. J. Innate Immun. 8, 374–385 (2016).
Vales-Gomez, M., Reyburn, H. T., Erskine, R. A., Lopez-Botet, M. & Strominger, J. L. Kinetics and peptide dependency of the binding of the inhibitory NK receptor CD94/NKG2-A and the activating receptor CD94/NKG2-C to HLA-E. EMBO J. 18, 4250–4260 (1999).
Hammer, Q., Ruckert, T. & Romagnani, C. Natural killer cell specificity for viral infections. Nat. Immunol. 19, 800–808 (2018).
Hammer, Q. et al. Peptide-specific recognition of human cytomegalovirus strains controls adaptive natural killer cells. Nat. Immunol. 19, 453–463 (2018).
Schlums, H. et al. Cytomegalovirus infection drives adaptive epigenetic diversification of NK cells with altered signaling and effector function. Immunity 42, 443–456 (2015).
Beziat, V. et al. CMV drives clonal expansion of NKG2C+ NK cells expressing self-specific KIRs in chronic hepatitis patients. Eur. J. Immunol. 42, 447–457 (2012).
Guma, M. et al. Human cytomegalovirus infection is associated with increased proportions of NK cells that express the CD94/NKG2C receptor in aviremic HIV-1-positive patients. J. Infect. Dis. 194, 38–41 (2006).
Lee, J. et al. Epigenetic modification and antibody-dependent expansion of memory-like NK cells in human cytomegalovirus-infected individuals. Immunity 42, 431–442 (2015).
Vivier, E. et al. Innate lymphoid cells: 10 years on. Cell 174, 1054–1066 (2018).
Scoville, S. D., Freud, A. G. & Caligiuri, M. A. Cellular pathways in the development of human and murine innate lymphoid cells. Curr. Opin. Immunol. 56, 100–106 (2019).
Freud, A. G., Mundy-Bosse, B. L., Yu, J. & Caligiuri, M. A. The broad spectrum of human natural killer cell diversity. Immunity 47, 820–833 (2017).
Bal, S. M., Golebski, K. & Spits, H. Plasticity of innate lymphoid cell subsets. Nat. Rev. Immunol. 20, 552–565 (2020).
Zhou, J., Tian, Z. & Peng, H. Tissue-resident NK cells and other innate lymphoid cells. Adv. Immunol. 145, 37–53 (2020).
Turner, J. E., Rickassel, C., Healy, H. & Kassianos, A. J. Natural killer cells in kidney health and disease. Front. Immunol. 10, 587 (2019).
Turner, J. E., Becker, M., Mittrucker, H. W. & Panzer, U. Tissue-resident lymphocytes in the kidney. J. Am. Soc. Nephrol. 29, 389–399 (2018).
Victorino, F. et al. Tissue-resident NK cells mediate ischemic kidney injury and are not depleted by anti-asialo-GM1 antibody. J. Immunol. 195, 4973–4985 (2015).
Parkes, M. D., Halloran, P. F. & Hidalgo, L. G. Evidence for CD16a-mediated NK cell stimulation in antibody-mediated kidney transplant rejection. Transplantation 101, e102–e111 (2017).
Apps, R. et al. HIV-1 Vpu mediates HLA-C downregulation. Cell Host Microbe 19, 686–695 (2016).
Ende, Z. et al. HLA class I downregulation by HIV-1 variants from subtype C transmission pairs. J. Virol. 92, e01633-17 (2018).
Lin, A., Xu, H. & Yan, W. Modulation of HLA expression in human cytomegalovirus immune evasion. Cell Mol. Immunol. 4, 91–98 (2007).
Aptsiauri, N., Ruiz-Cabello, F. & Garrido, F. The transition from HLA-I positive to HLA-I negative primary tumors: the road to escape from T-cell responses. Curr. Opin. Immunol. 51, 123–132 (2018).
Fangazio, M. et al. Genetic mechanisms of HLA-I loss and immune escape in diffuse large B cell lymphoma. Proc. Natl Acad. Sci. USA 118, e2104504118 (2021).
Puttick, C. et al. MHC Hammer reveals genetic and non-genetic HLA disruption in cancer evolution. Nat. Genet. 56, 2121–2131 (2024).
McGranahan, N. et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171, 1259–1271.e11 (2017).
Law, B. M. P. et al. Interferon-γ production by tubulointerstitial human CD56bright natural killer cells contributes to renal fibrosis and chronic kidney disease progression. Kidney Int. 92, 79–88 (2017).
Song, H. et al. Transforming growth factor-β1 regulates human renal proximal tubular epithelial cell susceptibility to natural killer cells via modulation of the NKG2D ligands. Int. J. Mol. Med. 36, 1180–1188 (2015).
Shinozaki, M. et al. IL-15, a survival factor for kidney epithelial cells, counteracts apoptosis and inflammation during nephritis. J. Clin. Invest. 109, 951–960 (2002).
Maddineni, S. et al. An intraepithelial ILC1-like natural killer cell subset produces IL-13. Front. Immunol. 16, 1521086 (2025).
Caligiuri, M. A. Human natural killer cells. Blood 112, 461–469 (2008).
Hadad, U., Martinez, O. & Krams, S. M. NK cells after transplantation: friend or foe. Immunol. Res. 58, 259–267 (2014).
Sagoo, P. et al. Development of a cross-platform biomarker signature to detect renal transplant tolerance in humans. J. Clin. Invest. 120, 1848–1861 (2010).
Ferlazzo, G. & Moretta, L. Dendritic cell editing by natural killer cells. Crit. Rev. Oncog. 19, 67–75 (2014).
Ferlazzo, G. et al. Human dendritic cells activate resting natural killer (NK) cells and are recognized via the NKp30 receptor by activated NK cells. J. Exp. Med. 195, 343–351 (2002).
Lv, D. et al. Advances in understanding of dendritic cell in the pathogenesis of acute kidney injury. Front. Immunol. 15, 1294807 (2024).
Cantoni, C. et al. Recent advances in the role of natural killer cells in acute kidney injury. Front. Immunol. 11, 1484 (2020).
Cao, Q., Harris, D. C. & Wang, Y. Macrophages in kidney injury, inflammation, and fibrosis. Physiology 30, 183–194 (2015).
Zhang, Z. X. et al. NK cells induce apoptosis in tubular epithelial cells and contribute to renal ischemia-reperfusion injury. J. Immunol. 181, 7489–7498 (2008).
Sadler, R. et al. The differential effect of Interferon-gamma on acute kidney injury and parasitemia in experimental malaria. Sci. Rep. 15, 6402 (2025).
Cyster, J. G. & Schwab, S. R. Sphingosine-1-phosphate and lymphocyte egress from lymphoid organs. Annu. Rev. Immunol. 30, 69–94 (2012).
Shannon, M. J. & Mace, E. M. Natural killer cell integrins and their functions in tissue residency. Front. Immunol. 12, 647358 (2021).
Kim, H. J. et al. TLR2 signaling in tubular epithelial cells regulates NK cell recruitment in kidney ischemia-reperfusion injury. J. Immunol. 191, 2657–2664 (2013).
Kim, H. J. et al. Reverse signaling through the costimulatory ligand CD137L in epithelial cells is essential for natural killer cell-mediated acute tissue inflammation. Proc. Natl Acad. Sci. USA 109, E13–E22 (2012).
Zhang, Z. X. et al. Osteopontin expressed in tubular epithelial cells regulates NK cell-mediated kidney ischemia reperfusion injury. J. Immunol. 185, 967–973 (2010).
Zou, X. et al. NK cell regulatory property is involved in the protective role of MSC-derived extracellular vesicles in renal ischemic reperfusion injury. Hum. Gene Ther. 27, 926–935 (2016).
Wu, I.-W. et al. Deep immune profiling of patients with renal impairment unveils distinct immunotypes associated with disease severity. Clin. Kidney J. 16, 78–89 (2022).
Burfeind, K. G., Funahashi, Y., Munhall, A. C., Eiwaz, M. & Hutchens, M. P. Natural killer lymphocytes mediate renal fibrosis due to acute cardiorenal syndrome. Kidney360 5, 8–21 (2024).
Furini, G. et al. Proteomic profiling reveals the transglutaminase-2 externalization pathway in kidneys after unilateral ureteric obstruction. J. Am. Soc. Nephrol. 29, 880–905 (2018).
Scarpellini, A. et al. Syndecan-4 knockout leads to reduced extracellular transglutaminase-2 and protects against tubulointerstitial fibrosis. J. Am. Soc. Nephrol. 25, 1013–1027 (2014).
Yadav, B., Prasad, N., Agrawal, V., Agarwal, V. & Jain, M. Lower circulating cytotoxic T-cell frequency and higher intragraft granzyme-B expression are associated with inflammatory interstitial fibrosis and tubular atrophy in renal allograft recipients. Medicina 59, 1175 (2023).
Rowshani, A. T. et al. Hyperexpression of the granzyme B inhibitor PI-9 in human renal allografts: a potential mechanism for stable renal function in patients with subclinical rejection. Kidney Int. 66, 1417–1422 (2004).
Choy, J. C. Granzymes and perforin in solid organ transplant rejection. Cell Death Differ. 17, 567–576 (2010).
Kuepper, M. et al. Increase in killer-specific secretory protein of 37 kDa in bronchoalveolar lavage fluid of allergen-challenged patients with atopic asthma. Clin. Exp. Allergy 35, 643–649 (2005).
Ogawa, K. et al. A novel serum protein that is selectively produced by cytotoxic lymphocytes. J. Immunol. 166, 6404–6412 (2001).
Maucourant, C. et al. Natural killer cell immunotypes related to COVID-19 disease severity. Sci. Immunol. 5, eabd6832 (2020).
Halloran, P. F. et al. Molecular diagnosis of ABMR with or without donor-specific antibody in kidney transplant biopsies: differences in timing and intensity but similar mechanisms and outcomes. Am. J. Transpl. 22, 1976–1991 (2022).
Halloran, P. F. et al. Review: the transcripts associated with organ allograft rejection. Am. J. Transpl. 18, 785–795 (2018).
Hidalgo, L. G. et al. NK cell transcripts and NK cells in kidney biopsies from patients with donor-specific antibodies: evidence for NK cell involvement in antibody-mediated rejection. Am. J. Transpl. 10, 1812–1822 (2010).
Taetzsch, T., Brayman, V. L. & Valdez, G. FGF binding proteins (FGFBPs): modulators of FGF signaling in the developing, adult, and stressed nervous system. Biochim. Biophys. Acta Mol. Basis Dis. 1864, 2983–2991 (2018).
Xie, Y. et al. FGF/FGFR signaling in health and disease. Signal Transduct. Target. Ther. 5, 181 (2020).
Hanneken, A., Maher, P. A. & Baird, A. High affinity immunoreactive FGF receptors in the extracellular matrix of vascular endothelial cells — implications for the modulation of FGF-2. J. Cell Biol. 128, 1221–1228 (1995).
Stauber, D. J., DiGabriele, A. D. & Hendrickson, W. A. Structural interactions of fibroblast growth factor receptor with its ligands. Proc. Natl Acad. Sci. USA 97, 49–54 (2000).
Hu, X. et al. Fibroblast growth factor 2 Is produced by renal tubular cells to act as a paracrine factor in maladaptive kidney repair after cisplatin nephrotoxicity. Lab. Invest. 103, 100009 (2023).
Livingston, M. J. et al. Autophagy activates EGR1 via MAPK/ERK to induce FGF2 in renal tubular cells for fibroblast activation and fibrosis during maladaptive kidney repair. Autophagy 20, 1032–1053 (2024).
Aggarwal, S. et al. SOX9 switch links regeneration to fibrosis at the single-cell level in mammalian kidneys. Science 383, eadd6371 (2024).
Robertson, M. J. Role of chemokines in the biology of natural killer cells. J. Leukoc. Biol. 71, 173–183 (2002).
Anders, H. J., Kitching, A. R., Leung, N. & Romagnani, P. Glomerulonephritis: immunopathogenesis and immunotherapy. Nat. Rev. Immunol. 23, 453–471 (2023).
Sylvestre, D. L. & Ravetch, J. V. Fc receptors initiate the arthus reaction: redefining the inflammatory cascade. Science 265, 1095–1098 (1994).
Spada, R. et al. NKG2D ligand overexpression in lupus nephritis correlates with increased NK cell activity and differentiation in kidneys but not in the periphery. J. Leukoc. Biol. 97, 583–598 (2015).
Naesens, M. et al. The Banff 2022 Kidney Meeting report: reappraisal of microvascular inflammation and the role of biopsy-based transcript diagnostics. Am. J. Transpl. 24, 338–349 (2024).
Reindl-Schwaighofer, R. et al. Contribution of non-HLA incompatibility between donor and recipient to kidney allograft survival: genome-wide analysis in a prospective cohort. Lancet 393, 910–917 (2019).
Sablik, M. et al. Microvascular inflammation of kidney allografts and clinical outcomes. N. Engl. J. Med. 392, 763–776 (2025).
Hirohashi, T. et al. A novel pathway of chronic allograft rejection mediated by NK cells and alloantibody. Am. J. Transpl. 12, 313–321 (2012).
Kohei, N. et al. Natural killer cells play a critical role in mediating inflammation and graft failure during antibody-mediated rejection of kidney allografts. Kidney Int. 89, 1293–1306 (2016).
Yagisawa, T. et al. In the absence of natural killer cell activation donor-specific antibody mediates chronic, but not acute, kidney allograft rejection. Kidney Int. 95, 350–362 (2019).
Miyairi, S. et al. Recipient myeloperoxidase-producing cells regulate antibody-mediated acute versus chronic kidney allograft rejection. JCI Insight 6, e148747 (2021).
Yazdani, S. et al. Natural killer cell infiltration is discriminative for antibody-mediated rejection and predicts outcome after kidney transplantation. Kidney Int. 95, 188–198 (2019).
Colvin, R. B. Antibody-mediated renal allograft rejection: diagnosis and pathogenesis. J. Am. Soc. Nephrol. 18, 1046–1056 (2007).
Mauiyyedi, S. et al. Chronic humoral rejection: identification of antibody-mediated chronic renal allograft rejection by C4d deposits in peritubular capillaries. J. Am. Soc. Nephrol. 12, 574–582 (2001).
Lamarthee, B. et al. Transcriptional and spatial profiling of the kidney allograft unravels a central role for FcyRIII+ innate immune cells in rejection. Nat. Commun. 14, 4359 (2023).
Bailly, E. et al. Interleukin-21 promotes Type-1 activation and cytotoxicity of CD56dimCD16bright natural killer cells during kidney allograft antibody-mediated rejection showing a new link between adaptive and innate humoral allo-immunity. Kidney Int. 104, 707–723 (2023).
Hill, P. A., Main, I. W. & Atkins, R. C. ICAM-1 and VCAM-1 in human renal allograft rejection. Kidney Int. 47, 1383–1391 (1995).
Alari-Pahissa, E. et al. Alloreactive adaptive natural killer cells in renal transplantation: potential contribution to allograft microvascular inflammation. Am. J. Transpl. 25, 1657–1669 (2025).
Diebold, M. et al. Natural killer cell presence in antibody-mediated rejection. Transpl. Int. 37, 13209 (2024).
Liu, L. L. et al. Critical Role of CD2 co-stimulation in adaptive natural killer cell responses revealed in NKG2C-deficient humans. Cell Rep. 15, 1088–1099 (2016).
Diebold, M. et al. Natural killer cell functional genetics and donor-specific antibody-triggered microvascular inflammation. Am. J. Transpl. 24, 743–754 (2024).
Redondo-Pachon, D. et al. Adaptive NKG2C+ NK cell response and the risk of cytomegalovirus infection in kidney transplant recipients. J. Immunol. 198, 94–101 (2017).
Llinas-Mallol, L. et al. Long-term redistribution of peripheral lymphocyte subpopulations after switching from calcineurin to mTOR inhibitors in kidney transplant recipients. J. Clin. Med. 9, 1088 (2020).
Hoffmann, U. et al. NK Cells of kidney transplant recipients display an activated phenotype that is influenced by immunosuppression and pathological staging. PLoS ONE 10, e0132484 (2015).
Neudoerfl, C. et al. The peripheral NK cell repertoire after kidney transplantation is modulated by different immunosuppressive drugs. Front. Immunol. 4, 46 (2013).
Bailly, E. et al. FCGR2C Q13 and FCGR3A V176 alleles jointly associate with worse natural killer cell-mediated antibody-dependent cellular cytotoxicity and microvascular inflammation in kidney allograft antibody-mediated rejection. Am. J. Transpl. 25, 302–315 (2025).
Diebold, M. et al. Effect of felzartamab on the molecular phenotype of antibody-mediated rejection in kidney transplant biopsies. Nat. Med. 31, 1668–1676 (2025).
Marks, W. H. et al. Safety and efficacy of eculizumab in the prevention of antibody-mediated rejection in living-donor kidney transplant recipients requiring desensitization therapy: a randomized trial. Am. J. Transpl. 19, 2876–2888 (2019).
Bhalla, A., Alachkar, N. & Alasfar, S. Complement-based therapy in the management of antibody-mediated rejection. Adv. Chronic Kidney Dis. 27, 138–148 (2020).
Tiller, G. et al. Weak expression of terminal complement in active antibody-mediated rejection of the kidney. Front. Immunol. 13, 845301 (2022).
Schmauch, E. et al. Integrative multi-omics profiling in human decedents receiving pig heart xenografts. Nat. Med. 30, 1448–1460 (2024).
Loupy, A. et al. Immune response after pig-to-human kidney xenotransplantation: a multimodal phenotyping study. Lancet 402, 1158–1169 (2023).
Uehara, S., Chase, C. M., Colvin, R. B., Russell, P. S. & Madsen, J. C. Further evidence that NK cells may contribute to the development of cardiac allograft vasculopathy. Transpl. Proc. 37, 70–71 (2005).
Uehara, S. et al. NK cells can trigger allograft vasculopathy: the role of hybrid resistance in solid organ allografts. J. Immunol. 175, 3424–3430 (2005).
Koenig, A. et al. Missing self triggers NK cell-mediated chronic vascular rejection of solid organ transplants. Nat. Commun. 10, 5350 (2019).
Callemeyn, J. et al. Missing self-induced microvascular rejection of kidney allografts: a population-based study. J. Am. Soc. Nephrol. 32, 2070–2082 (2021).
Callemeyn, J. et al. Transcriptional changes in kidney allografts with histology of antibody-mediated rejection without anti-HLA donor-specific antibodies. J. Am. Soc. Nephrol. 31, 2168–2183 (2020).
Diebold, M. et al. Functional natural killer-cell genetics and microvascular inflammation after kidney transplantation: an observational cohort study. Transplantation 109, 860–870 (2025).
Koenig, A. et al. Missing self-induced activation of NK cells combines with non-complement-fixing donor-specific antibodies to accelerate kidney transplant loss in chronic antibody-mediated rejection. J. Am. Soc. Nephrol. 32, 479–494 (2021).
Bohmig, G. A., Loupy, A., Sablik, M. & Naesens, M. Microvascular inflammation in kidney allografts: new directions for patient management. Am. J. Transpl. 25, 1410–1416 (2025).
Ruan, D. F. et al. High-dimensional analysis of NK cells in kidney transplantation uncovers subsets associated with antibody-independent graft dysfunction. JCI Insight 9, e185687 (2024).
Lin, C. M., Gill, R. G. & Mehrad, B. The natural killer cell activating receptor, NKG2D, is critical to antibody-dependent chronic rejection in heart transplantation. Am. J. Transpl. 21, 3550–3560 (2021).
Maier, S. et al. Inhibition of natural killer cells results in acceptance of cardiac allografts in CD28−/− mice. Nat. Med. 7, 557–562 (2001).
Diebold, M. et al. Chronic rejection after kidney transplantation. Transplantation 109, 610–621 (2025).
Mayer, K. A. et al. A randomized phase 2 trial of felzartamab in antibody-mediated rejection. N. Engl. J. Med. 391, 122–132 (2024).
Haustein, S. et al. Interleukin-15 receptor blockade in non-human primate kidney transplantation. Transplantation 89, 937–944 (2010).
Shin, B. H. et al. Regulation of anti-HLA antibody-dependent natural killer cell activation by immunosuppressive agents. Transplantation 97, 294–300 (2014).
Ashraf, M. I. et al. Natural killer cells promote kidney graft rejection independently of cyclosporine A therapy. Front. Immunol. 10, 2279 (2019).
Raulet, D. H. & Vance, R. E. Self-tolerance of natural killer cells. Nat. Rev. Immunol. 6, 520–531 (2006).
Acknowledgements
The authors thank R. Fairchild for critical review of the manuscript before submission. Additionally, the authors thank A. Freud for personal discussion on NK cell development and differentiation pathways. The work was partly supported by NIH grant U01 AI 70424 to P.H.
Author information
Authors and Affiliations
Contributions
Both authors researched data for the article, made substantial contributions to discussions of the content and wrote, reviewed or edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Nephrology thanks Georg Böhmig, Luis Hidalgo and Maarten Naesens for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Horowitz, A., Heeger, P. Natural killer cells in kidney immune surveillance, injury and fibrosis. Nat Rev Nephrol (2025). https://doi.org/10.1038/s41581-025-01029-x
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41581-025-01029-x