Abstract
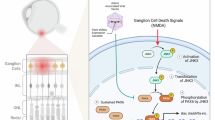
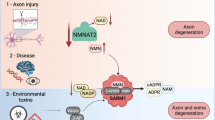
Programmed axon degeneration (PAxD) is an evolutionarily conserved mechanism in the nervous system that is activated by axonal injury (axotomy) to execute the self-destruction of a severed distal axon. It can also be triggered by non-axotomy insults, resulting in the loss of axons connected to their cell bodies. PAxD is therefore a promising target for therapeutic intervention and drugs that inhibit it are currently being tested in clinical trials. In this Review, we summarize the molecular mechanism of PAxD, focusing on its regulation by nicotinamide adenine dinucleotide (NAD+) metabolism and how it dictates Ca2+-mediated axonal demise. We examine its involvement in human disease and its potential as a therapeutic target by dissecting its role in various non-axotomy disease models. Finally, we address key challenges for its clinical translation, including the need for relevant biomarkers and safety considerations. Further advancements in understanding PAxD will pave the way for new therapeutic strategies targeting human axonopathies.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Neukomm, L. J. & Freeman, M. R. Diverse cellular and molecular modes of axon degeneration. Trends Cell Biol. 24, 515–523 (2014).
Burgess, R. W. & Crish, S. D. Editorial: axonopathy in neurodegenerative disease. Front. Neurosci. 12, 769 (2018).
Waller, A. Experiments on the section of the glossopharyngeal and hypoglossal nerves of the frog, and observations of the alterations produced thereby in the structure of their primitive fibres. Phil. Trans. R. Soc. Lond. 140, 423–429 (1850).
Llobet Rosell, A. & Neukomm, L. J. Axon death signalling in Wallerian degeneration among species and in disease. Open Biol. 9, 190118 (2019).
Figley, M. D. & DiAntonio, A. The SARM1 axon degeneration pathway: control of the NAD+ metabolome regulates axon survival in health and disease. Curr. Opin. Neurobiol. 63, 59–66 (2020).
Sambashivan, S. & Freeman, M. R. SARM1 signaling mechanisms in the injured nervous system. Curr. Opin. Neurobiol. 69, 247–255 (2021).
Coleman, M. P. & Höke, A. Programmed axon degeneration: from mouse to mechanism to medicine. Nat. Rev. Neurosci. 21, 183–196 (2020).
Waller, T. J. & Collins, C. A. Multifaceted roles of SARM1 in axon degeneration and signaling. Front. Cell Neurosci. 16, 958900 (2022).
Alexandris, A. & Koliatsos, V. E. NAD+, axonal maintenance, and neurological disease. Antioxid. Redox Signal. 39, 1167–1184 (2023).
McGuinness, H. Y., Gu, W., Shi, Y., Kobe, B. & Ve, T. SARM1-dependent axon degeneration: nucleotide signaling, neurodegenerative disorders, toxicity, and therapeutic opportunities. Neuroscientist 30, 473–492 (2024).
Loreto, A., Antoniou, C., Merlini, E., Gilley, J. & Coleman, M. P. NMN: the NAD precursor at the intersection between axon degeneration and anti-ageing therapies. Neurosci. Res. 197, 18–24 (2023).
Cao, Y., Wang, Y. & Yang, J. NAD+-dependent mechanism of pathological axon degeneration. Cell Insight 1, 100019 (2022).
Alberti, C. et al. Charcot-Marie-tooth disease type 2A: an update on pathogenesis and therapeutic perspectives. Neurobiol. Dis. 193, 106467 (2024).
Loreto, A., Merlini, E. & Coleman, M. P. Programmed axon death: a promising target for treating retinal and optic nerve disorders. Eye 38, 1802–1809 (2024).
Tarasiuk, O., Molteni, L., Malacrida, A. & Nicolini, G. The role of NMNAT2/SARM1 in neuropathy development. Biology 13, 61 (2024).
Park, S. B. et al. Axonal degeneration in chemotherapy-induced peripheral neurotoxicity: clinical and experimental evidence. J. Neurol. Neurosurg. Psychiatry 94, 962–972 (2023).
Merlini, E., Coleman, M. P. & Loreto, A. Mitochondrial dysfunction as a trigger of programmed axon death. Trends Neurosci. 45, 53–63 (2022).
Krauss, R., Bosanac, T., Devraj, R., Engber, T. & Hughes, R. O. Axons matter: the promise of treating neurodegenerative disorders by targeting SARM1-Mediated axonal degeneration. Trends Pharmacol. Sci. 41, 281–293 (2020).
Conforti, L., Gilley, J. & Coleman, M. P. Wallerian degeneration: an emerging axon death pathway linking injury and disease. Nat. Rev. Neurosci. 15, 394–409 (2014).
Di Stefano, M. et al. A rise in NAD precursor nicotinamide mononucleotide (NMN) after injury promotes axon degeneration. Cell Death Differ. 22, 731–742 (2015). This study demonstrates that NMN accumulates after axotomy and its rise drives axon degeneration.
Jiang, Y. et al. The NAD+-mediated self-inhibition mechanism of pro-neurodegenerative SARM1. Nature 33, 245–246 (2020). This article reveals the cryo-electron microscopy structure of the full-length SARM1 protein and identified an NAD+-mediated self-inhibitory mechanism via binding to the ARM domain.
Figley, M. D. et al. SARM1 is a metabolic sensor activated by an increased NMN/NAD+ ratio to trigger axon degeneration. Neuron 109, 1118–1136.e11 (2021). This study demonstrates that SARM1, an inducible pro-degenerative NADase, is a metabolic sensor activated by an increase in the NMN-to-NAD+ ratio.
Mori, V. et al. Metabolic profiling of alternative NAD biosynthetic routes in mouse tissues. PLoS ONE 9, e113939 (2014).
Gilley, J., Orsomando, G., Nascimento-Ferreira, I. & Coleman, M. P. Absence of SARM1 rescues development and survival of NMNAT2-deficient axons. Cell Rep. 10, 1974–1981 (2015). This report reveals that axon degeneration specifically induced by NMNAT2 depletion requires SARM1, suggesting a linear NMNAT2–SARM1 signalling pathway.
Sasaki, Y., Nakagawa, T., Mao, X., DiAntonio, A. & Milbrandt, J. NMNAT1 inhibits axon degeneration via blockade of SARM1-mediated NAD+ depletion. eLife 5, 1010 (2016).
Llobet Rosell, A. et al. The NAD+ precursor NMN activates dSarm to trigger axon degeneration in Drosophila. eLife 11, e80245 (2022).
Imai, S.-I. Nicotinamide phosphoribosyltransferase (Nampt): a link between NAD biology, metabolism, and diseases. Curr. Pharm. Des. 15, 20–28 (2009).
Ratajczak, J. et al. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 7, 13103 (2016).
Sasaki, Y., Araki, T. & Milbrandt, J. Stimulation of nicotinamide adenine dinucleotide biosynthetic pathways delays axonal degeneration after axotomy. J. Neurosci. 26, 8484–8491 (2006).
Sasaki, Y. et al. Nicotinic acid mononucleotide is an allosteric SARM1 inhibitor promoting axonal protection. Exp. Neurol. 345, 113842 (2021).
Berger, F., Lau, C., Dahlmann, M. & Ziegler, M. Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J. Biol. Chem. 280, 36334–36341 (2005).
Yan, T. et al. Nmnat2 delays axon degeneration in superior cervical ganglia dependent on its NAD synthesis activity. Neurochem. Int. 56, 101–106 (2010).
Summers, D. W., Milbrandt, J. & DiAntonio, A. Palmitoylation enables MAPK-dependent proteostasis of axon survival factors. Proc. Natl Acad. Sci. USA 11, E8746–E8754 (2018).
Yang, J. et al. Pathological axonal death through a MAPK cascade that triggers a local energy deficit. Cell 160, 161–176 (2015).
Walker, L. J. et al. MAPK signaling promotes axonal degeneration by speeding the turnover of the axonal maintenance factor NMNAT2. eLife 6, 545 (2017).
Miller, B. R. et al. A dual leucine kinase-dependent axon self-destruction program promotes Wallerian degeneration. Nat. Neurosci. 12, 387–389 (2009).
Shin, J. E. et al. SCG10 is a JNK target in the axonal degeneration pathway. Proc. Natl Acad. Sci. USA 109, E3696–E3705 (2012).
Gilley, J. & Coleman, M. P. Endogenous Nmnat2 is an essential survival factor for maintenance of healthy axons. PLoS Biol. 8, e1000300 (2010). This article reveals endogenous NMNAT2 as a labile axon survival factor whose constant replenishment by anterograde axonal transport is a limiting factor for axon survival.
Xiong, X. et al. The highwire ubiquitin ligase promotes axonal degeneration by tuning levels of Nmnat protein. PLoS Biol. 10, e1001440 (2012). This study reveals that the evolutionarily conserved E3 ubiquitin ligase Highwire in Drosophila promotes axon degeneration by inducing the rapid degradation of the Nmnat protein.
Babetto, E., Beirowski, B., Russler, E. V., Milbrandt, J. & DiAntonio, A. The Phr1 ubiquitin ligase promotes injury-induced axon self-destruction. Cell Rep. 3, 1422–1429 (2013). This article demonstrates that the mammalian E3 ubiquitin ligase MYCBP2 (also known as PHR1) promotes the rapid degradation of NMNAT2 in axons of the peripheral and central nervous system.
Neukomm, L. J., Burdett, T. C., Gonzalez, M. A., Zuchner, S. & Freeman, M. R. Rapid in vivo forward genetic approach for identifying axon death genes in Drosophila. Proc. Natl Acad. Sci. USA 111, 9965–9970 (2014).
Desbois, M. et al. PAM forms an atypical SCF ubiquitin ligase complex that ubiquitinates and degrades NMNAT2. J. Biol. Chem. 293, 13897–13909 (2018).
Yamagishi, Y. & Tessier-Lavigne, M. An atypical SCF-like ubiquitin ligase complex promotes wallerian degeneration through regulation of axonal Nmnat2. Cell Rep. 17, 774–782 (2016). This study reveals that the E3 ubiquitin ligase component SKP1A regulates protein levels of NMNAT2 in axons.
Milde, S., Gilley, J. & Coleman, M. P. Subcellular localization determines the stability and axon protective capacity of axon survival factor nmnat2. PLoS Biol. 11, e1001539 (2013).
Milde, S., Fox, A. N., Freeman, M. R. & Coleman, M. P. Deletions within its subcellular targeting domain enhance the axon protective capacity of Nmnat2 in vivo. Sci. Rep. 3, 2567 (2013).
Osterloh, J. M. et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481–484 (2012). This report identifies Sarm in Drosophila and SARM1 in mice as a executioner of axotomy-induced PAxD.
Gerdts, J., Summers, D. W., Sasaki, Y., DiAntonio, A. & Milbrandt, J. Sarm1-mediated axon degeneration requires both SAM and TIR interactions. J. Neurosci. 33, 13569–13580 (2013).
Gerdts, J., Brace, E. J., Sasaki, Y., DiAntonio, A. & Milbrandt, J. SARM1 activation triggers axon degeneration locally via NAD+ destruction. Science 348, 453–457 (2015). This study reveals that the dimerization of the SARM1 TIR domains initiates a rapid axonal breakdown of NAD+ after axotomy.
Essuman, K. et al. The SARM1 Toll/interleukin-1 receptor domain possesses intrinsic NAD+ cleavage activity that promotes pathological axonal degeneration. Neuron 93, 1334–1343.e5 (2017). This article demonstrates that TIR domains of SARM1 have intrinsic NADase activity and deplete axonal NAD+ to induce pathological axon loss.
Neukomm, L. J. et al. Axon death pathways converge on axundead to promote functional and structural axon disassembly. Neuron 95, 78–91.e5 (2017). This study identifies Axundead as required for axotomy-induced axon degeneration downstream of Sarm and Nmnat in Drosophila.
Sporny, M. et al. Structural evidence for an octameric ring arrangement of SARM1. J. Mol. Biol. 431, 3591–3605 (2019).
Horsefield, S. et al. NAD+ cleavage activity by animal and plant TIR domains in cell death pathways. Science 365, 793–799 (2019). This article reveals that NAD+ cleavage in the octameric SARM1 structure is mediated by TIR domain self-association both in animals and plants.
Bratkowski, M. et al. Structural and mechanistic regulation of the pro-degenerative NAD hydrolase SARM1. Cell Rep. 32, 107999 (2020). This report reveals mechanistic insights into the regulation of SARM1 activity by revealing cryo-electron microscopy structures of autoinhibited and activated SARM1.
Sporny, M. et al. Structural basis for SARM1 inhibition and activation under energetic stress. eLife 9, W344 (2020).
Shen, C. et al. Multiple domain interfaces mediate SARM1 autoinhibition. Proc. Natl Acad. Sci. USA 118, e2023151118 (2021).
Shi, Y. et al. Structural basis of SARM1 activation, substrate recognition, and inhibition by small molecules. Mol. Cell 82, 1643–1659.e10 (2022). This study demonstrates that a base-exchange reaction underlies potent orthosteric inhibition of SARM1 by a series of isoquinoline compounds.
Angeletti, C. et al. SARM1 is a multi-functional NAD(P)ase with prominent base exchange activity, all regulated bymultiple physiologically relevant NAD metabolites. iScience 25, 103812 (2022).
Essuman, K. et al. TIR domain proteins are an ancient family of NAD+-consuming enzymes. Curr. Biol. 28, 421–430.e4 (2018).
Parrish, A. B., Freel, C. D. & Kornbluth, S. Cellular mechanisms controlling caspase activation and function. Cold Spring Harb. Perspect. Biol. 5, a008672 (2013).
Zhao, Z. Y. et al. A cell-permeant mimetic of NMN activates SARM1 to produce cyclic ADP-ribose and induce non-apoptotic cell death. iScience 15, 452–466 (2019). This article demonstrates that CZ-48, a cell-permeant mimetic of NMN, activates SARM1 to induce non-apoptotic cell death.
Li, W. H. et al. Permeant fluorescent probes visualize the activation of SARM1 and uncover an anti-neurodegenerative drug candidate. eLife 10, e67381 (2021). This report identifies PC6 as a substrate of SARM1 that undergoes a large red fluorescent shift upon conversion into PAD6, enabling its use as a SARM1 activity reporter.
George, E. B., Glass, J. D. & Griffin, J. W. Axotomy-induced axonal degeneration is mediated by calcium influx through ion-specific channels. J. Neurosci. 15, 6445–6452 (1995).
Schlaepfer, W. W. Calcium-induced degeneration of axoplasm in isolated segments of rat peripheral nerve. Brain Res. 69, 203–215 (1974).
Schlaepfer, W. W. Structural alterations of peripheral nerve induced by the calcium ionophore A23187. Brain Res. 136, 1–9 (1977).
Knöferle, J. et al. Mechanisms of acute axonal degeneration in the optic nerve in vivo. Proc. Natl Acad. Sci. USA 107, 6064–6069 (2010).
Adalbert, R. et al. Intra-axonal calcium changes after axotomy in wild-type and slow wallerian degeneration axons. Neuroscience 225, 44–54 (2012).
Avery, M. A. et al. WldS prevents axon degeneration through increased mitochondrial flux and enhanced mitochondrial Ca2+ buffering. Curr. Biol. 22, 596–600 (2012).
Vargas, M. E., Yamagishi, Y., Tessier-Lavigne, M. & Sagasti, A. Live imaging of calcium dynamics during axon degeneration reveals two functionally distinct phases of calcium influx. J. Neurosci. 35, 15026–15038 (2015).
Mishra, B., Carson, R., Hume, R. I. & Collins, C. A. Sodium and potassium currents influence wallerian degeneration of injured Drosophila axons. J. Neurosci. 33, 18728–18739 (2013).
Villegas, R. et al. Calcium release from intra-axonal endoplasmic reticulum leads to axon degeneration through mitochondrial dysfunction. J. Neurosci. 34, 7179–7189 (2014).
Loreto, A., Di Stefano, M., Gering, M. & Conforti, L. Wallerian degeneration is executed by an NMN-SARM1-dependent late Ca2+ influx but only modestly influenced by mitochondria. Cell Rep. 13, 2539–2552 (2015).
Ko, K. W., DeVault, L., Sasaki, Y., Milbrandt, J. & DiAntonio, A. Live imaging reveals the cellular events downstream of SARM1 activation. eLife 10, e71148 (2021).
Guse, A. H. Calcium mobilizing second messengers derived from NAD. Biochim. Biophys. Acta. 1854, 1132–1137 (2015).
Sasaki, Y. et al. cADPR is a gene dosage-sensitive biomarker of SARM1 activity in healthy, compromised, and degenerating axons. Exp. Neurol. 329, 113252 (2020). This study reveals that cADPR, a product of SARM1-dependent cleavage of NAD+, serves as a biomarker in cultured neurons, sciatic nerve and the brain in preclinical models.
Garb, J. et al. The SARM1 TIR domain produces glycocyclic ADPR molecules as minor products. PLoS ONE 19, e0302251 (2024).
Blomgren, K. Calpastatin is upregulated and acts as a suicide substrate to calpains in neonatal rat hypoxia‐ischemia. Ann. NY Acad. Sci. 890, 270–271 (1999).
Yang, J. et al. Regulation of axon degeneration after injury and in development by the endogenous calpain inhibitor calpastatin. Neuron 80, 1175–1189 (2013).
Ma, M. et al. Calpains mediate axonal cytoskeleton disintegration during Wallerian degeneration. Neurobiol. Dis. 56, 34–46 (2013).
Macqueen, D. J. & Wilcox, A. H. Characterization of the definitive classical calpain family of vertebrates using phylogenetic, evolutionary and expression analyses. Open Biol. 4, 130219 (2014).
Bridge, P. M. et al. Nerve crush injuries — a model for axonotmesis. Exp. Neurol. 127, 284–290 (1994).
Maxwell, W. L., Bartlett, E. & Morgan, H. Wallerian degeneration in the optic nerve stretch-injury model of traumatic brain injury: a stereological analysis. J. Neurotrauma 32, 780–790 (2015).
Geisler, S. Augustus Waller’s foresight realized: SARM1 in peripheral neuropathies. Curr. Opin. Neurobiol. 87, 102884 (2024).
Griffin, J. W. et al. Macrophage responses and myelin clearance during Wallerian degeneration: relevance to immune-mediated demyelination. J. Neuroimmunol. 40, 153–165 (1992).
Vaquié, A. et al. Injured axons instruct schwann cells to build constricting actin spheres to accelerate axonal disintegration. Cell Rep. 27, 3152–3166.e7 (2019).
MacDonald, J. M. et al. The Drosophila cell corpse engulfment receptor draper mediates glial clearance of severed axons. Neuron 50, 869–881 (2006).
Sasaki, Y. & Milbrandt, J. Axonal degeneration is blocked by nicotinamide mononucleotide adenylyltransferase (Nmnat) protein transduction into transected axons. J. Biol. Chem. 285, 41211–41215 (2010).
Hughes, R. O. et al. Small molecule SARM1 inhibitors recapitulate the SARM1–/– phenotype and allow recovery of a metastable pool of axons fated to degenerate. Cell Rep. 34, 108588 (2021). This report describes a potent SMI of SARM1.
Mack, T. G. et al. Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat. Neurosci. 4, 1199–1206 (2001). This article reports that the Ube4b/Nmnat chimeric gene is necessary and sufficient to protect injured axons.
Lunn, E. R., Perry, V. H., Brown, M. C., Rosen, H. & Gordon, S. Absence of Wallerian degeneration does not hinder regeneration in peripheral nerve. Eur. J. Neurosci. 1, 27–33 (1989).
Paglione, M. et al. Local translatome sustains synaptic function in impaired Wallerian degeneration. EMBO Rep. 26, 61–83 (2024).
Sasaki, Y., Vohra, B. P. S., Lund, F. E. & Milbrandt, J. Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J. Neurosci. 29, 5525–5535 (2009).
Sasaki, Y., Vohra, B. P. S., Baloh, R. H. & Milbrandt, J. Transgenic mice expressing the Nmnat1 protein manifest robust delay in axonal degeneration in vivo. J. Neurosci. 29, 6526–6534 (2009).
Feng, Y. et al. Overexpression of WldS or Nmnat2 in mauthner cells by single-cell electroporation delays axon degeneration in live Zebrafish. J. Neurosci. Res. 88, 3319–3327 (2010).
Avery, M. A., Sheehan, A. E., Kerr, K. S., Wang, J. & Freeman, M. R. Wld S requires Nmnat1 enzymatic activity and N16-VCP interactions to suppress Wallerian degeneration. J. Cell Biol. 184, 501–513 (2009).
Babetto, E. et al. Targeting NMNAT1 to axons and synapses transforms its neuroprotective potency in vivo. J. Neurosci. 30, 13291–13304 (2010).
Paglione, M., Llobet Rosell, A., Chatton, J.-Y. & Neukomm, L. J. Morphological and functional evaluation of axons and their synapses during axon death in Drosophila melanogaster. J. Vis. Exp. https://doi.org/10.3791/60865 (2020).
Fang, Y., Soares, L., Teng, X., Geary, M. & Bonini, N. M. A novel Drosophila model of nerve injury reveals an essential role of Nmnat in maintaining axonal integrity. Curr. Biol. 22, 590–595 (2012).
Di Stefano, M. et al. NMN deamidase delays Wallerian degeneration and rescues axonal defects caused by NMNAT2 deficiency in vivo. Curr. Biol. 27, 784–794 (2017).
Gould, S. A. et al. Sarm1 haploinsufficiency or low expression levels after antisense oligonucleotides delay programmed axon degeneration. Cell Rep. 37, 110108 (2021).
Liu, P. et al. Differential effects of SARM1 inhibition in traumatic glaucoma and EAE optic neuropathies. Mol. Ther. Nucleic Acids 32, 13–27 (2023).
Loreto, A. et al. SARM1 activation induces reversible mitochondrial dysfunction and can be prevented in human neurons by antisense oligonucleotides. Neurobiol. Dis. 213, 106986 (2025).
Geisler, S. et al. Gene therapy targeting SARM1 blocks pathological axon degeneration in mice. J. Exp. Med. 216, 294–303 (2019).
Bratkowski, M. et al. Uncompetitive, adduct-forming SARM1 inhibitors are neuroprotective in preclinical models of nerve injury and disease. Neuron 110, 3711–3726.e16 (2022). This study reveals that BEIs are highly potent and confer compelling neuroprotection in pre-clinical models of neurological injury and disease.
Leahey, R. R. et al. Therapeutic safety implications of SARM1 active site inhibitors: subinhibitory concentrations cause neurodegeneration. npj Drug Discov. 2, 21 (2025). This study shows that subinhibitory concentrations of SARM1 BEIs, under mildly SARM1-activating conditions, paradoxically cause sustained SARM1 activation and toxicity.
Giroud, M. et al. Discovery of a potent SARM1 base-exchange inhibitor with in vivo efficacy. J. Med. Chem. 68, 6558–6575 (2025).
Mani, A. et al. SARM1 base-exchange inhibitors induce SARM1 activation and neurodegeneration at low doses. npj Drug Discov. 2, 12 (2025).
Bosanac, T. et al. Pharmacological SARM1 inhibition protects axon structure and function in paclitaxel-induced peripheral neuropathy. Brain 44, 3226–3238 (2021). This report demonstrates that potent and selective irreversible isothiazole inhibitors of SARM1 enzymatic activity protect axons in a mouse model of chemotherapy-induced peripheral neuropathy.
Loring, H. S., Parelkar, S. S., Mondal, S. & Thompson, P. R. Identification of the first noncompetitive SARM1 inhibitors. Bioorganic Med. Chem. 28, 115644 (2020).
Feldman, H. C. et al. Selective inhibitors of SARM1 targeting an allosteric cysteine in the autoregulatory ARM domain. Proc. Natl Acad. Sci. USA 119, e2208457119 (2022).
Khazma, T. et al. A duplex structure of SARM1 octamers stabilized by a new inhibitor. Cell Mol. Life Sci. 80, 16 (2023).
Schlaepfer, W. W. & Hasler, M. B. Characterization of the calcium-induced disruption of neurofilaments in rat peripheral nerve. Brain Res. 168, 299–309 (1979).
Jayaram, H. N., Kusumanchi, P. & Yalowitz, J. A. NMNAT expression and its relation to NAD metabolism. Curr. Med. Chem. 18, 1962–1972 (2011).
Yamamoto, M. et al. Nmnat3 is dispensable in mitochondrial NAD level maintenance in vivo. PLoS ONE 11, e0147037 (2016).
Gilley, J., Adalbert, R., Yu, G. & Coleman, M. P. Rescue of peripheral and CNS axon defects in mice lacking NMNAT2. J. Neurosci. 33, 13410–13424 (2013).
Loreto, A. et al. Mitochondrial impairment activates the Wallerian pathway through depletion of NMNAT2 leading to SARM1-dependent axon degeneration. Neurobiol. Dis. 134, 104678 (2019).
Geisler, S. et al. Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. JCI Insight 4, e129920 (2019).
Sasaki, Y. et al. SARM1 depletion rescues NMNAT1-dependent photoreceptor cell death and retinal degeneration. eLife 9, 817 (2020).
Grozio, A. et al. Slc12a8 is a nicotinamide mononucleotide transporter. Nat. Metab. 1, 47–57 (2019).
Schmidt, M. S. & Brenner, C. Absence of evidence that Slc12a8 encodes a nicotinamide mononucleotide transporter. Nat. Metab. 1, 660–661 (2019).
Grozio, A. et al. Reply to: absence of evidence that Slc12a8 encodes a nicotinamide mononucleotide transporter. Nat. Metab. 1, 662–665 (2019).
Loreto, A. et al. Neurotoxin-mediated potent activation of the axon degeneration regulator SARM1. eLife 10, e72823 (2021). This study reveals that the neurotoxin vacor, metabolized by NAMPT into the NMN analogue VMN, also acts as a potent SARM1 activator.
Wu, T. et al. Neurotoxins subvert the allosteric activation mechanism of SARM1 to induce neuronal loss. Cell Rep. 37, 109872 (2021).
Huang, Y. et al. Stepwise activation of SARM1 for cell death and axon degeneration revealed by a biosynthetic NMN mimic. Proc. Natl. Acad. Sci. USA 122, e2424906122 (2025).
Gilley, J. et al. Enrichment of SARM1 alleles encoding variants with constitutively hyperactive NADase in patients with ALS and other motor nerve disorders. eLife 10, e70905 (2021).
Bloom, A. J. et al. Constitutively active SARM1 variants that induce neuropathy are enriched in ALS patients. Mol. Neurodegener. 17, 1 (2022).
Zhao, Y. J. et al. Acidic pH irreversibly activates the signaling enzyme SARM1. FEBS J. 288, 6783–6794 (2021).
Loring, H. S. et al. A phase transition enhances the catalytic activity of SARM1, an NAD+ glycohydrolase involved in neurodegeneration. eLife 10, e66694 (2021).
Icso, J. D. & Thompson, P. R. A phase transition reduces the threshold for nicotinamide mononucleotide-based activation of SARM1, an NAD(P) hydrolase, to physiologically relevant levels. J. Biol. Chem. 299, 105284 (2023).
Tribble, J. R. et al. NMNAT2 is a druggable target to drive neuronal NAD production. Nat. Commun. 15, 6256 (2024).
Fang, F. et al. NMNAT2 is downregulated in glaucomatous RGCs and RGC-specific gene therapy rescues neurodegeneration and visual function. Mol. Ther. 30, 1421–1431 (2022).
US National Library of Medicine. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT05492201 (2022). The phase I clinical trial using SARM1 SMIs in healthy participants.
Australian New Zealand ClinicalTrials Registry. ANZCTR https://anzctr.org.au/Trial/Registration/TrialReview.aspx?id=385567 (2024). Successfully completed phase I clinical trial using SARM1 SMIs in healthy volunteers.
Coleman, M. P. Axon biology in ALS: mechanisms of axon degeneration and prospects for therapy. Neurotherapeutics 19, 1133–1144 (2022).
Moss, K. R. & Höke, A. Targeting the programmed axon degeneration pathway as a potential therapeutic for Charcot-Marie-Tooth disease. Brain Res. 1727, 146539 (2020).
Herbosa, C. G., Perez, R., Jaeger, A., Dy, C. J. & Brogan, D. M. Inhibition of SARM1 reduces neuropathic pain in a spared nerve injury rodent model. Muscle Nerve 71, 670–679 (2025).
Lukacs, M. et al. Severe biallelic loss-of-function mutations in nicotinamide mononucleotide adenylyltransferase 2 (NMNAT2) in two fetuses with fetal akinesia deformation sequence. Exp. Neurol. 320, 112961 (2019). This report reveals severe biallelic LOF mutations in human NMNAT2.
Hicks, A. N., Campeau, L., Burmeister, D., Bishop, C. E. & Andersson, K. Lack of nicotinamide mononucleotide adenylyltransferase 2 (Nmnat2): consequences for mouse bladder development and function. Neurourol. Urodyn. 32, 1130–1136 (2013).
Dingwall, C. B. et al. Macrophage depletion blocks congenital SARM1-dependent neuropathy. J. Clin. Invest. 132, e159800 (2022).
Huppke, P. et al. Homozygous NMNAT2 mutation in sisters with polyneuropathy and erythromelalgia. Exp. Neurol. 320, 112958 (2019). The study reveals homozygous partial LOF mutations in human NMNAT2.
Chiang, P.-W. et al. Exome sequencing identifies NMNAT1 mutations as a cause of Leber congenital amaurosis. Nat. Genet. 44, 972–974 (2012).
Falk, M. J. et al. NMNAT1 mutations cause Leber congenital amaurosis. Nat. Genet. 44, 1040–1045 (2012).
Koenekoop, R. K. et al. Mutations in NMNAT1 cause Leber congenital amaurosis and identify a new disease pathway for retinal degeneration. Nat. Genet. 44, 1035–1039 (2012).
Perrault, I. et al. Mutations in NMNAT1 cause Leber congenital amaurosis with early-onset severe macular and optic atrophy. Nat. Genet. 44, 975–977 (2012).
Yi, Z. et al. Clinical features and genetic spectrum of NMNAT1-associated retinal degeneration. Eye 36, 2279–2285 (2022).
Kumaran, N., Robson, A. G. & Michaelides, M. A novel case series of NMNAT1-associated early-onset retinal dystrophy: extending the phenotypic spectrum. Retin. Cases Brief Rep. 15, 139–144 (2021).
Sasaki, Y., Margolin, Z., Borgo, B., Havranek, J. J. & Milbrandt, J. Characterization of Leber congenital amaurosis-associated NMNAT1 mutants*. J. Biol. Chem. 290, 17228–17238 (2015).
Ademi, M., Yang, X., Coleman, M. P. & Gilley, J. Natural variants of human SARM1 cause both intrinsic and dominant loss-of-function influencing axon survival. Sci. Rep. 12, 13846 (2022).
van Rheenen, W. et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat. Genet. 48, 1043–1048 (2016).
Fogh, I. et al. A genome-wide association meta-analysis identifies a novel locus at 17q11.2 associated with sporadic amyotrophic lateral sclerosis. Hum. Mol. Genet. 23, 2220–2231 (2014).
LeWitt, P. A. The neurotoxicity of the rat poison vacor — a clinical study of 12 cases. N. Engl. J. Med. 302, 73–77 (1980).
Ling, Y. et al. The expanding role of pyridine and dihydropyridine scaffolds in drug design. Drug Des. Dev. Ther. 15, 4289–4338 (2021).
Antenor-Dorsey, J. A. V. & O’Malley, K. L. WldS but not Nmnat1 protects dopaminergic neurites from MPP+ neurotoxicity. Mol. Neurodegener. 7, 5 (2012).
Brazill, J. M., Cruz, B., Zhu, Y. & Zhai, R. G. Nmnat mitigates sensory dysfunction in a Drosophila model of paclitaxel-induced peripheral neuropathy. Dis. Model. Mech. 11, dmm032938 (2018).
Press, C. & Milbrandt, J. Nmnat delays axonal degeneration caused by mitochondrial and oxidative stress. J. Neurosci. 28, 4861–4871 (2008).
Wang, M. S. et al. The Wld S protein protects against axonal degeneration: a model of gene therapy for peripheral neuropathy. Ann. Neurol. 50, 773–779 (2001).
Wang, M.-S., Wu, Y., Culver, D. G. & Glass, J. D. The gene for slow Wallerian degeneration (Wlds) is also protective against vincristine neuropathy. Neurobiol. Dis. 8, 155–161 (2001).
Geisler, S. et al. Prevention of vincristine-induced peripheral neuropathy by genetic deletion of SARM1 in mice. Brain 139, 3092–3108 (2016).
Summers, D. W., Gibson, D. A., DiAntonio, A. & Milbrandt, J. SARM1-specific motifs in the TIR domain enable NAD+ loss and regulate injury-induced SARM1 activation. Proc. Natl Acad. Sci. USA 113, E6271–E6280 (2016).
Turkiew, E., Falconer, D., Reed, N. & Höke, A. Deletion of Sarm1 gene is neuroprotective in two models of peripheral neuropathy. J. Peripher. Nerv. Syst. 22, 162–171 (2017).
Gilley, J., Mayer, P. R., Yu, G. & Coleman, M. P. Low levels of NMNAT2 compromise axon development and survival. Hum. Mol. Genet. 28, 448–458 (2019).
Li, Y. et al. Sarm1 activation produces cADPR to increase intra-axonal Ca++ and promote axon degeneration in PIPN. J. Cell Biol. 221, e202106080 (2022).
Hinz, F. I. et al. Context-specific stress causes compartmentalized SARM1 activation and local degeneration in cortical neurons. J. Neurosci. 44, e2424232024 (2024).
Chen, Y.-H., Sasaki, Y., DiAntonio, A. & Milbrandt, J. SARM1 is required in human derived sensory neurons for injury-induced and neurotoxic axon degeneration. Exp. Neurol. 339, 113636 (2021).
Gomez-Deza, J., Slavutsky, A. L., Nebiyou, M. & Pichon, C. E. L. Local production of reactive oxygen species drives vincristine-induced axon degeneration. Cell Death Dis. 14, 807 (2023).
Cetinkaya-Fisgin, A. et al. Cisplatin induced neurotoxicity is mediated by Sarm1 and calpain activation. Sci. Rep. 10, 21889 (2020).
Gould, S. A. et al. Protection against oxaliplatin-induced mechanical and thermal hypersensitivity in Sarm1–/– mice. Exp. Neurol. 338, 113607 (2021).
Snavely, A. R. et al. Bortezomib-induced neurotoxicity in human neurons is the consequence of nicotinamide adenine dinucleotide depletion. Dis. Model. Mech. 15, dmm049358 (2022).
Klemmensen, M. M., Borrowman, S. H., Pearce, C., Pyles, B. & Chandra, B. Mitochondrial dysfunction in neurodegenerative disorders. Neurotherapeutics 21, e00292 (2024).
Yamada, Y. et al. A SARM1/mitochondrial feedback loop drives neuropathogenesis in a Charcot-Marie-Tooth disease type 2A rat model. J. Clin. Invest. 132, e161566 (2022).
Peters, O. M. et al. Loss of Sarm1 does not suppress motor neuron degeneration in the SOD1G93A mouse model of amyotrophic lateral sclerosis. Hum. Mol. Genet. 11, 3761–3771 (2018).
Collins, J. M. et al. Sarm1 knockout modifies biomarkers of neurodegeneration and spinal cord circuitry but not disease progression in the mSOD1G93A mouse model of ALS. Neurobiol. Dis. 172, 105821 (2022).
Velde, C. V., Garcia, M. L., Yin, X., Trapp, B. D. & Cleveland, D. W. The neuroprotective factor WldS does not attenuate mutant SOD1-mediated motor neuron disease. Neuromolecular Med. 5, 193–203 (2004).
Fischer, L. R. et al. The WldS gene modestly prolongs survival in the SOD1G93A fALS mouse. Neurobiol. Dis. 19, 293–300 (2005).
White, M. A. et al. Sarm1 deletion suppresses TDP-43-linked motor neuron degeneration and cortical spine loss. Acta Neuropathol. Commun. 7, 166 (2019).
Scotter, E. L., Chen, H.-J. & Shaw, C. E. TDP-43 proteinopathy and ALS: insights into disease mechanisms and therapeutic targets. Neurotherapeutics 12, 352–363 (2015).
Peters, O. M. et al. Genetic diversity of axon degenerative mechanisms in models of Parkinson’s disease. Neurobiol. Dis. 155, 105368 (2021).
Antoniou, C. et al. Chronically low NMNAT2 expression causes Sub-lethal SARM1 activation and altered response to nicotinamide riboside in axons. Mol. Neurobiol. 62, 3903–3917 (2024).
Sun, Y. et al. Sarm1-mediated neurodegeneration within the enteric nervous system protects against local inflammation of the colon. Protein Cell 12, 621–638 (2021).
He, S. et al. Targeting SARM1 as a novel neuroprotective therapy in neurotropic viral infections. J. Neuroinflammation 22, 113 (2025).
Sundaramoorthy, V. et al. Novel role of SARM1 mediated axonal degeneration in the pathogenesis of rabies. PLoS Pathog. 16, e1008343 (2020). This study demonstrates that SARM1 is necessary for the rapid progression of rabies-induced axonal degeneration and that it hinders the spread of rabies virus among connected neurons.
Xu, G. et al. PARP-1 mediated cell death is directly activated by ZIKV infection. Virology 537, 254–262 (2019).
Pang, H. et al. Aberrant NAD+ metabolism underlies Zika virus-induced microcephaly. Nat. Metab. 3, 1109–1124 (2021).
Crawford, C. L. et al. SARM1 depletion slows axon degeneration in a CNS model of neurotropic viral infection. Front. Mol. Neurosci. 15, 860410 (2022).
Szretter, K. J. et al. The immune adaptor molecule SARM modulates tumor necrosis factor alpha production and microglia activation in the brainstem and restricts West Nile virus pathogenesis. J. Virol. 83, 9329–9338 (2009).
Mukherjee, P., Woods, T. A., Moore, R. A. & Peterson, K. E. Activation of the innate signaling molecule MAVS by bunyavirus infection upregulates the adaptor protein SARM1, leading to neuronal death. Immunity 38, 705–716 (2013).
Hou, Y.-J. et al. SARM is required for neuronal injury and cytokine production in response to central nervous system viral infection. J. Immunol. 191, 875–883 (2013).
Uccellini, M. B. et al. Passenger mutations confound phenotypes of SARM1-deficient mice. Cell Rep. 31, 107498 (2020).
Rouse, B. T. & Sehrawat, S. Immunity and immunopathology to viruses: what decides the outcome? Nat. Rev. Immunol. 10, 514–526 (2010).
Zhu, C., Li, B., Frontzek, K., Liu, Y. & Aguzzi, A. SARM1 deficiency up-regulates XAF1, promotes neuronal apoptosis, and accelerates prion disease. J. Exp. Med. 216, 743–756 (2019).
Xiang, L. et al. SARM1 deletion in parvalbumin neurons is associated with autism-like behaviors in mice. Cell Death Dis. 13, 638 (2022).
Izadifar, A. et al. Axon morphogenesis and maintenance require an evolutionary conserved safeguard function of Wnk kinases antagonizing Sarm and Axed. Neuron 109, 2864–2883.e8 (2021).
Wu, C., Wairkar, Y. P., Collins, C. A. & DiAntonio, A. Highwire function at the Drosophila neuromuscular junction: spatial, structural, and temporal requirements. J. Neurosci. 25, 9557–9566 (2005).
Wan, H. I. et al. Highwire regulates synaptic growth in Drosophila. Neuron 26, 313–329 (2000).
Chen, C.-Y., Lin, C.-W., Chang, C.-Y., Jiang, S.-T. & Hsueh, Y.-P. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J. Cell Biol. 193, 769–784 (2011).
Lin, C.-W., Liu, H.-Y., Chen, C.-Y. & Hsueh, Y.-P. Neuronally-expressed Sarm1 regulates expression of inflammatory and antiviral cytokines in brains. Innate Immun. 20, 161–172 (2013).
Lin, C.-W. & Hsueh, Y.-P. Sarm1, a neuronal inflammatory regulator, controls social interaction, associative memory and cognitive flexibility in mice. Brain Behav. Immun. 37, 142–151 (2014).
Lin, C.-W., Chen, C.-Y., Cheng, S.-J., Hu, H.-T. & Hsueh, Y.-P. Sarm1 deficiency impairs synaptic function and leads to behavioral deficits, which can be ameliorated by an mGluR allosteric modulator. Front. Cell. Neurosci. 8, 87 (2014).
Li, W., Zhu, W., Chen, J., Ali, T. & Li, S. SARM1 deficiency induced depressive-like behavior via AMPKα/p-eEF2 axis to synapse dysfunction. Neuropharmacology 262, 110206 (2025).
Green, S. A. et al. Optimization of brain penetrant SARM1 orthosteric inhibitors and discovery of their paradoxical subinhibitory activation. ACS Med. Chem. Lett. 16, 1147–1154 (2025).
Reardon, H. T. et al. Base exchange inhibitors of SARM1 form mononucleotide adducts and activate SARM1 in vivo. Preprint at bioRxiv https://doi.org/10.1101/2025.04.22.649875 (2025).
Zhang, W. et al. SARM1 activation promotes axonal degeneration via a two-step phase transition. Nat. Chem. Biol. https://doi.org/10.1038/s41589-025-02009-9 (2025). This article reveals that SARM1 BEIs form covalent inhibitor–ADPR conjugates within TIR dimers, acting as molecular glues to promote superhelical TIR filament-mediated SARM1 assemblies that condense into stable, phase-separated aggregates with full NADase activity.
Murata, H. et al. c-Jun N-terminal kinase (JNK)-mediated phosphorylation of SARM1 regulates NAD+ cleavage activity to inhibit mitochondrial respiration. J. Biol. Chem. 293, 18933–18943 (2018).
Murata, H. et al. Phosphorylated SARM1 is involved in the pathological process of rotenone-induced neurodegeneration. J. Biochem. 174, 533–548 (2023).
Hopkins, E. L., Gu, W., Kobe, B. & Coleman, M. P. A novel NAD signaling mechanism in axon degeneration and its relationship to innate immunity. Front. Mol. Biosci. 8, 703532 (2021).
Avetisyan, A., Barria, R., Sheehan, A. & Freeman, M. R. An ionic sensor acts in parallel to dsarm to promote neurodegeneration. Preprint at bioRxiv https://doi.org/10.1101/2024.10.29.620922 (2024).
Zhu, W. J. et al. Gap junction intercellular communications regulates activation of SARM1 and protects against axonal degeneration. Cell Death Dis. 16, 13 (2025).
Dingwall, C. B. et al. Suppressing phagocyte activation by overexpressing the phosphatidylserine lipase ABHD12 preserves sarmopathic nerves. iScience 28, 112626 (2025).
Babetto, E., Wong, K. M. & Beirowski, B. A glycolytic shift in Schwann cells supports injured axons. Nat. Neurosci. 23, 1215–1228 (2020).
Mutschler, C. et al. Schwann cells are axo-protective after injury irrespective of myelination status in mouse Schwann cell/neuron cocultures. J. Cell Sci. 136, jcs261557 (2023).
Mariano, V., Domínguez-Iturza, N., Neukomm, L. J. & Bagni, C. Maintenance mechanisms of circuit-integrated axons. Curr. Opin. Neurobiol. 53, 162–173 (2018).
Salvadores, N., Sanhueza, M., Manque, P. & Court, F. A. Axonal degeneration during aging and its functional role in neurodegenerative disorders. Front. Neurosci. 11, 451 (2017).
Faust, T. E., Gunner, G. & Schafer, D. P. Mechanisms governing activity-dependent synaptic pruning in the developing mammalian CNS. Nat. Rev. Neurosci. 22, 657–673 (2021).
Furusawa, K. & Emoto, K. Spatiotemporal regulation of developmental neurite pruning: molecular and cellular insights from Drosophila models. Neurosci. Res. 167, 54–63 (2021).
Riccomagno, M. M. & Kolodkin, A. L. Sculpting neural circuits by axon and dendrite pruning. Annu. Rev. Cell Dev. Biol. 31, 779–805 (2015).
Schuldiner, O. & Yaron, A. Mechanisms of developmental neurite pruning. Cell. Mol. Life Sci. 72, 101–119 (2015).
Knoerl, R. et al. Exploring clinical markers of Axon degeneration processes in chemotherapy-induced peripheral neuropathy among young adults receiving vincristine or paclitaxel. BMC Neurol. 24, 366 (2024).
Khalil, M. et al. Neurofilaments as biomarkers in neurological disorders. Nat. Rev. Neurol. 14, 577–589 (2018).
Khalil, M. et al. Neurofilaments as biomarkers in neurological disorders — towards clinical application. Nat. Rev. Neurol. 20, 269–287 (2024).
Rosengren, L. E., Karlsson, J., Karlsson, J., Persson, L. I. & Wikkelsø, C. Patients with amyotrophic lateral sclerosis and other neurodegenerative diseases have increased levels of neurofilament protein in CSF. J. Neurochem. 67, 2013–2018 (1996).
Gisslén, M. et al. Plasma concentration of the Neurofilament Light protein (NFL) is a biomarker of CNS injury in HIV infection: a cross-sectional study. EBioMedicine 3, 135–140 (2016).
Kuhle, J. et al. Comparison of three analytical platforms for quantification of the neurofilament light chain in blood samples: ELISA, electrochemiluminescence immunoassay and Simoa. Clin. Chem. Lab. Med. 54, 1655–1661 (2016).
Rissin, D. M. et al. Single-molecule enzyme-linked immunosorbent assay detects serum proteins at subfemtomolar concentrations. Nat. Biotechnol. 28, 595–599 (2010).
Fazal, S. V. et al. SARM1 detection in myelinating glia: sarm1/Sarm1 is dispensable for PNS and CNS myelination in zebrafish and mice. Front. Cell. Neurosci. 17, 1158388 (2023).
Huang, K. et al. Base-exchange enabling the visualization of SARM1 activities in sciatic nerve-injured mice. ACS Sens. 8, 767–773 (2023).
Acknowledgements
The authors thank A. DiAntonio, A. Höke, B. Kobe, G. Orsomando, J. Gilley, M. Coleman and Y. Sasaki for their constructive feedback on the manuscript. A.L. is supported by the Faculty of Medicine and Health and Save Sight Institute New Academic Staff Funding (University of Sydney) and the Snow Vision Accelerator (Snow Medical). L.J.N. is supported by two Swiss National Science Foundation (SNSF) Assistant Professor starting grants (PP00P3_176855 and PP00P3_211015), a SNSF project grant (320030-236186) and the University of Lausanne/Canton de Vaud.
Author information
Authors and Affiliations
Contributions
The authors both contributed to all aspects of the manuscript preparation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Neuroscience thanks the anonymous reviewers for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Antisense oligonucleotides
-
Short synthetic strands of DNA or RNA designed to bind to specific target RNA molecules to block the RNA from producing a protein, altering its normal function or stability.
- Axonopathies
-
Neurological disorders characterized by progressive axonal degeneration, leading to disrupted neuronal communication.
- Base-exchange activity
-
The ability of an enzyme to swap one base for another within a larger molecule, often a nicotinamide adenine dinucleotide (NAD+)-related (or phospholipid) compound.
- Base-exchange inhibitors
-
(BEIs). They work as a base within the catalytic site of sterile-α and TIR motif-containing protein 1 (SARM1) by replacing nicotinamide in the NAD+ substrate to form a stable base–adenosine diphosphate-ribose adduct, thereby blocking SARM1 NADase activity.
- Glycohydrolase
-
An enzyme that breaks glycosidic bonds — using either water (hydrolysis) or alternative nucleophiles — thereby degrading complex substrates into simpler components.
- Passenger mutations
-
In the context of genetically modified organisms, passenger mutations are unintended genetic differences from a donor’s genetic background that are inadvertently transferred to a recipient during the genetic engineering process, potentially causing experimental bias or an unexpected phenotype.
- Phase transition
-
Reversible or irreversible changes in the physical state of proteins from liquid-like condensates into solid aggregates altering organization and function.
- Transglycosidase
-
An enzyme whose activity transfers a glycosyl (sugar) group from one molecule to another rather than hydrolysing it.
- Wallerian degeneration
-
The process of axotomy-induced distal axon degeneration and the clearance of the resulting debris by surrounding glia and immune cells, named after August Waller.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Loreto, A., Neukomm, L.J. Programmed axon degeneration: mechanism, inhibition and therapeutic potential. Nat. Rev. Neurosci. 27, 44–60 (2026). https://doi.org/10.1038/s41583-025-00986-3
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41583-025-00986-3