Fig. 1: Two distinct clades related to type V Cas12a2 nucleases exhibit RNA-activated cleavage of RNA but not DNA.
From: RNA-triggered Cas12a3 cleaves tRNA tails to execute bacterial immunity
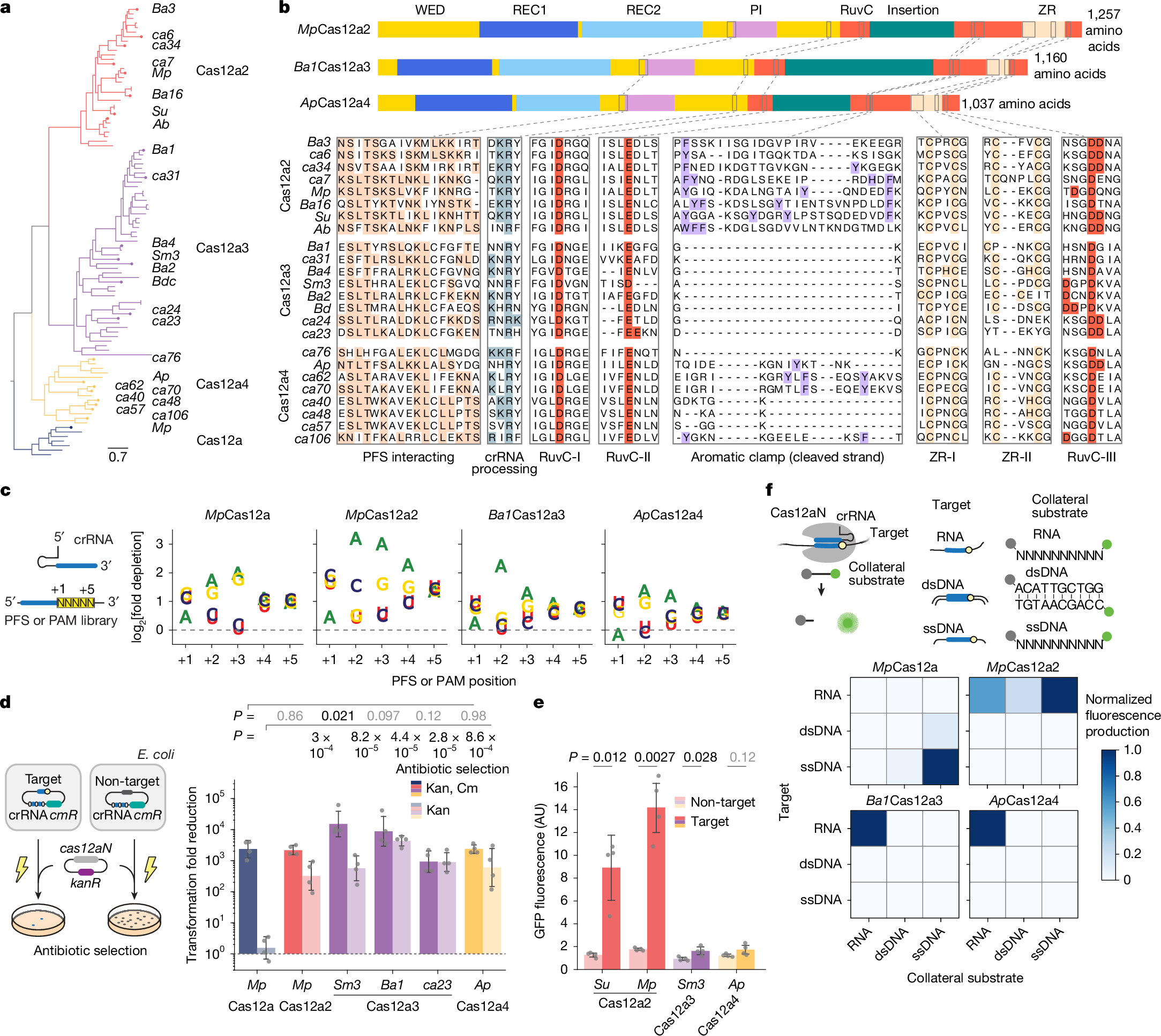
a, Phylogenetic analysis depicting Cas12a and the related Cas12a2, Cas12a3 and Cas12a4 clades. b, Domain and targeted sequence alignment across representative Cas12a2, Cas12a3 and Cas12a4 nucleases. c, Schematic (left) and quantification (right) of the nucleotide-depletion screen as part of the PFS (Cas12a2, Cas12a3 and Cas12a4) or protospacer-adjacent motif (PAM; Cas12a) determination for representative nucleases. Additional PFS screens are presented in Supplementary Fig. 2a. Results are the averages of independent experiments (n = 2). Note that the PAM for MpCas12a is the reverse complement of the sequence commonly reported for Cas12a nucleases (YYV). d, Schematic (left) and quantification (right) of the assessment of plasmid clearance versus growth arrest in E. coli based on variations of a plasmid interference assay. Plasmid clearance and growth arrest were differentiated on the basis of antibiotic selection for the target plasmid or the target plasmid and the nuclease plasmid. cmR, chloramphenicol-resistance cassette; kanR, kanamycin resistance cassette; Kan, kanamycin; Cm, chloramphenicol. e, Quantification of induction of the SOS DNA damage response in E. coli based on a transcriptional fluorescent reporter. Bars and error bars in d and e represent the geometric mean ± geometric s.d. and the mean ± s.d., respectively, of independent experiments starting from separate colonies (n = 4), with grey dots representing each measurement. f, Schematic (top) and measurement (bottom) of the assessment of different targets and cleavage substrates in vitro. Values represent the mean of independent experiments (n = 3 or 4). Complete time courses are shown in Fig. 2a and Supplementary Fig. 2e. Statistical analyses were performed using two-tailed Welch’s t-tests with all biological replicates. P values that are not significant (P ≥ 0.05) are shown in grey. AU, arbitrary units. Illustrations of thunderbolts and Petri dishes in d reproduced from ref. 27, CC BY 4.0.
