Fig. 2: Cas12a3 members preferentially cleave the conserved tail of tRNAs.
From: RNA-triggered Cas12a3 cleaves tRNA tails to execute bacterial immunity
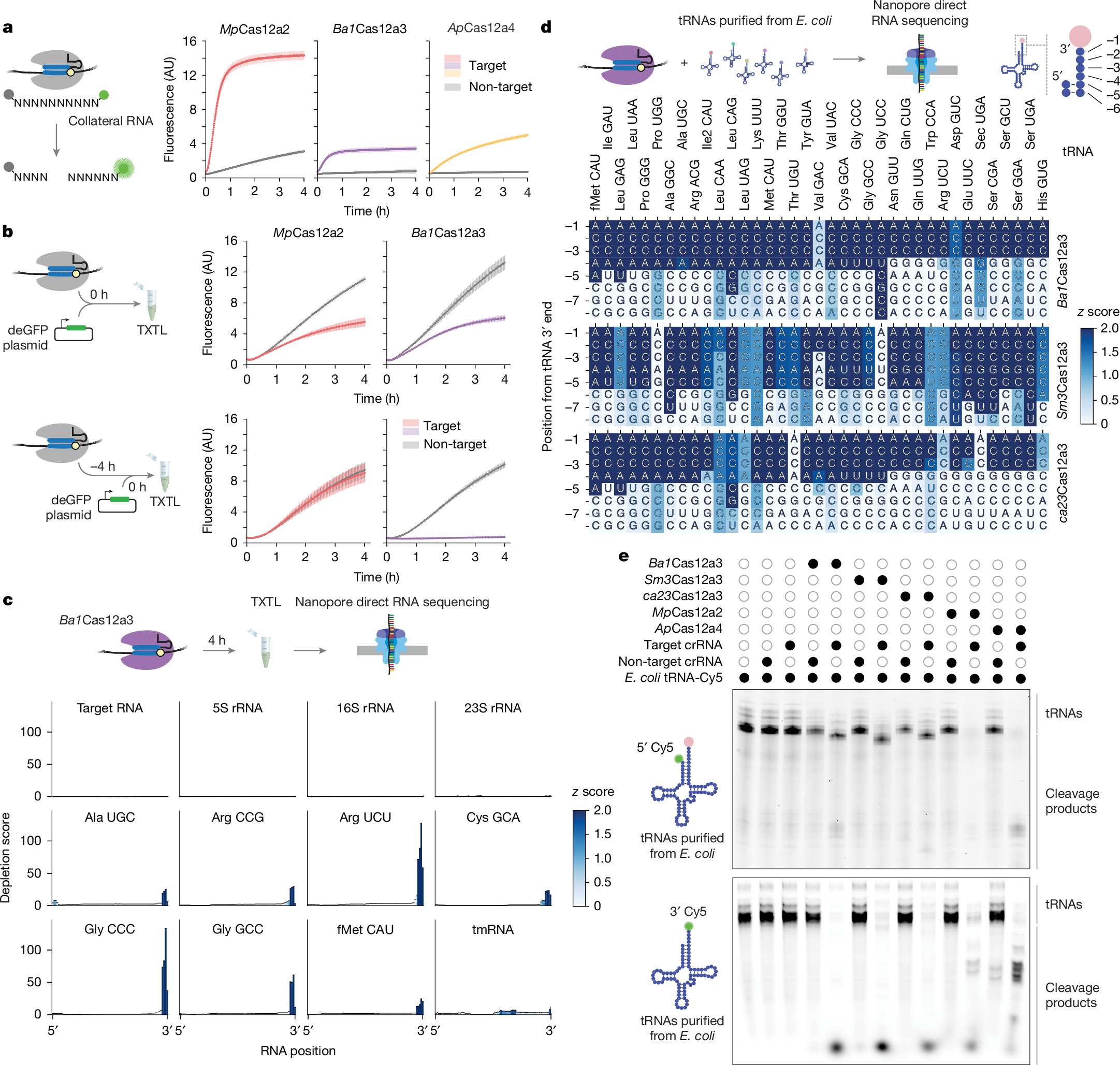
a, Schematic (left) and quantification (right) of the time course of cleavage of the RNA substrate library from Fig. 1f. Dots and bars represent the mean ± s.d. from independently mixed in vitro reactions (n = 4). AU, arbitrary units. b, Schematic (left) and quantification (right) of the fluorescence time courses of TXTL assays assessing nuclease activity based on silencing of deGFP expression. Dots and bars represent the mean and s.d. from independently mixed TXTL reactions (n = 3 or 4). c, Schematic (top) and quantification (bottom) of RNA sequencing of total RNA ≤ 200 nucleotides by Nanopore from TXTL reactions with Ba1Cas12a3 and a target or non-target RNA after 4 h. Values represent independent TXTL reactions (n = 3). The colour map indicates the z score of depletion scores at each position. d, Schematic (top) and quantification (bottom) of Nanopore sequencing of purified E. coli MRE600 tRNAs incubated with Ba1Cas12a3, Sm3Cas12a3 or ca23Cas12a3 under targeting versus non-targeting conditions. Values represent independent reactions (n = 3), with depletion z scores for each nucleotide as a colour map. Shown tRNAs (labelled with the corresponding anticodons) were detected in every nuclease reaction across the three independent experiments. Apparent cleavage sites ending in A could be shorter by one or (in the case of tRNAfMet(CAU)) two nucleotides towards the 3′ end owing to the use of a poly(A) extension to the cleavage product. See Supplementary Figs. 3 and 5 for full tRNA sequences with single-nucleotide depletion scores. e, Cleavage patterns of E. coli MRE600 tRNAs incubated with activated nucleases. The tRNA pool was 5′ labelled with a fluorophore, leaving the amino acyl group attached (top), or by replacing the 3′ amino acyl group with a fluorophore (bottom). Gel images are representative of independent cleavage reactions (n = 3). Open and closed circles represent the absence or presence, respectively, of the indicated component. For gel source data, see Supplementary Fig. 1.
