Fig. 1: Germline and somatic instability of common CAG-repeat alleles.
From: Insights into DNA repeat expansions among 900,000 biobank participants
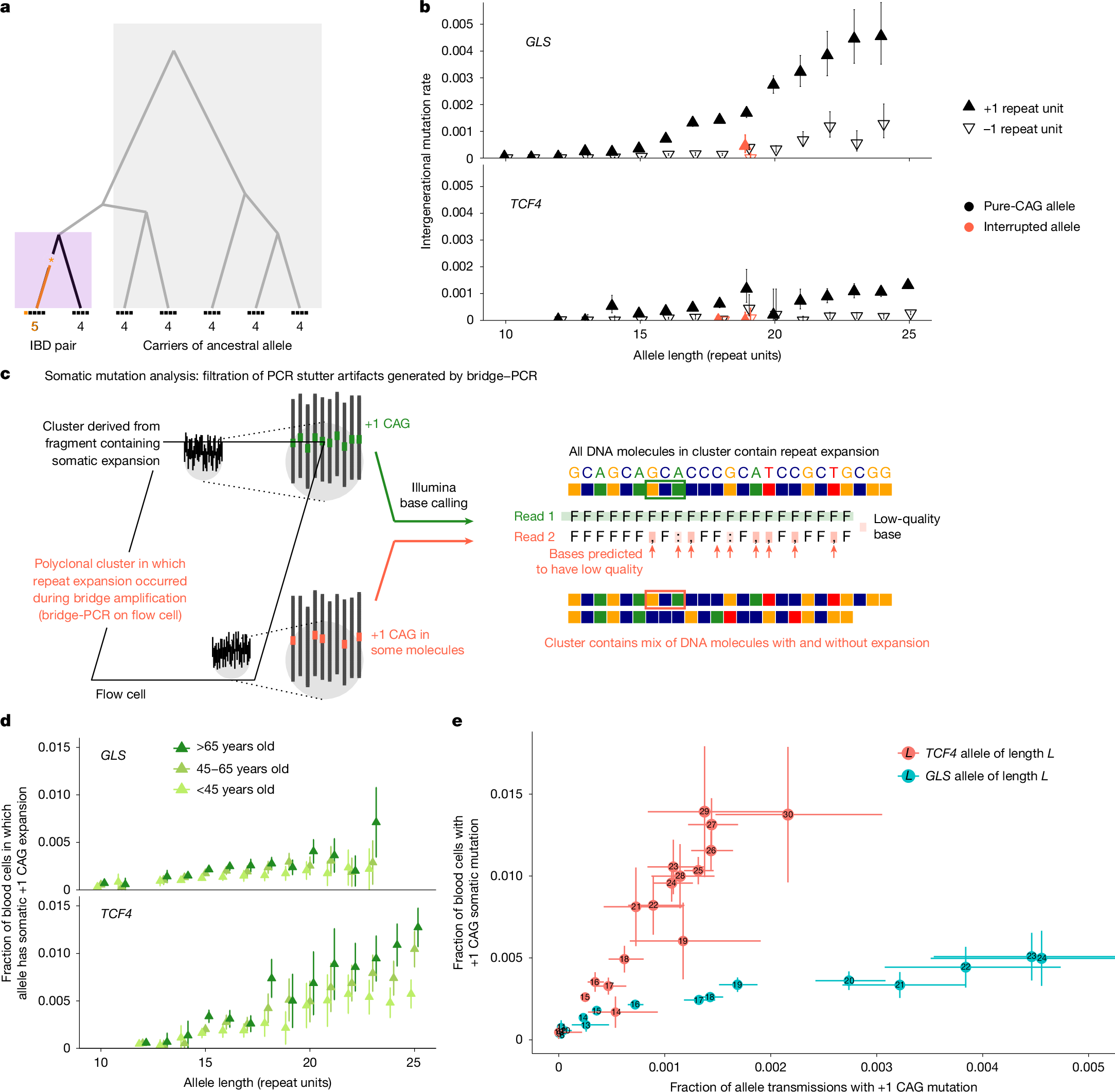
a, Germline mutation rates were estimated by analysing discordance rates among alleles inherited within IBD tracts shared by pairs of UKB participants. Ancestral alleles were imputed from more-distantly shared haplotypes. b, Per-generation rates of germline expansion (+1 repeat unit) and contraction (−1 repeat unit) of GLS and TCF4 repeat alleles, estimated in the UKB. c, The analytical strategy for estimating somatic mutation rates by detecting and filtering out reads that are likely to reflect PCR artifacts introduced during sequencing. During PCR-based bridge amplification on a flow cell, a DNA fragment is clonally amplified into a cluster of colocalized DNA molecules. A PCR stutter error results in a polyclonal cluster containing a mixture of DNA molecules with and without the error. If the molecules containing the error constitute the majority of the cluster, the sequencing read generated from the cluster (reflecting the majority base at each position within the read) will contain the error, but the heterogeneity of the cluster will reduce base qualities at positions within the read that mismatch between molecules with and without the error. d, The rates of somatic expansion of GLS and TCF4 repeat alleles (that is, the fractions of blood cells in which an allele has expanded by +1 repeat unit), stratified by age in AoU. e, Somatic mutation rates in the UKB plotted against germline mutation rates for GLS and TCF4 repeat alleles. The error bars show the 95% confidence intervals (CIs). Sample sizes are provided in Supplementary Table 3.
