Fig. 1: Retrospective quantification of EBV DNA in the UKB.
From: Population-scale sequencing resolves determinants of persistent EBV DNA
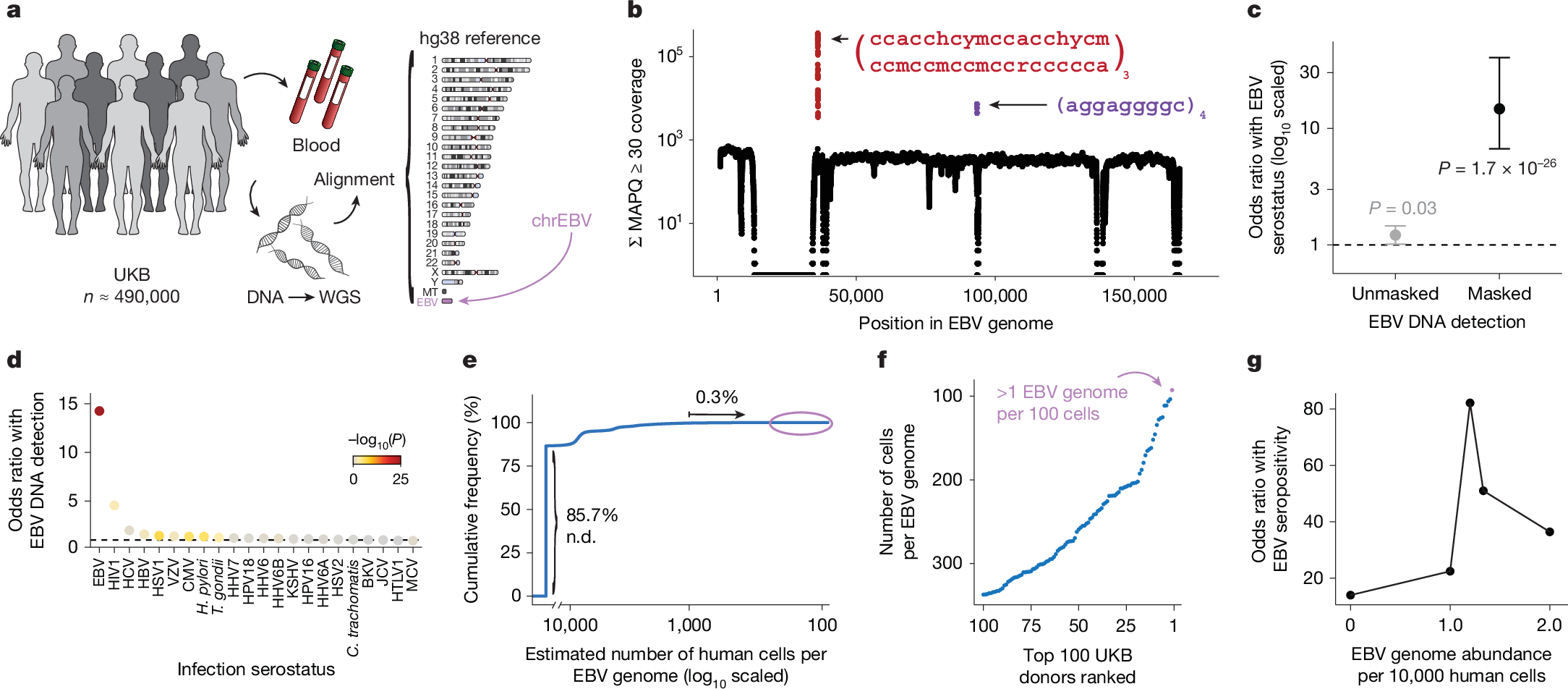
a, Schematic of the approach. WGS libraries from peripheral blood were aligned to the hg38 reference genome, which contains an EBV reference contig (chrEBV). Reads mapping to chrEBV were extracted for downstream analyses. b, Sum of per-base read coverage of high-confidence EBV-mapping reads. Two repetitive regions with inflated coverage are noted in red and purple (following IUPAC convention: h = A/C/T, y = C/T, m = A/C, r = A/G; subscripts indicate the number of repeats). c, Association summary of individual-level serostatus and EBV DNA quantification with variable region masking. Statistical test: two-sided Fisher’s exact test. Error bars represent 95% confidence intervals for the point effect estimate (centre dot). d, Summary of EBV DNA detection with serostatus of 22 infectious agents. Statistical test: two-sided Fisher’s exact test. HHV-6 was partitioned into strains HHV-6A and HHV-6B. e, Empirical cumulative distribution of detected EBV DNA across the entire cohort (85.7% of individuals had no detectable (n.d.) EBV DNA; 0.3% had EBV DNA at a copy number of 1+ EBV genome per 1,000 human cells). f, Top 100 individuals on the basis of EBV DNA copy number, from the circled population in e. g, Association between EBV seropositivity and EBV DNA detection thresholds at variable levels. Statistical test: two-sided Fisher’s exact test. Sample size of full UKB cohort: n = 490,560. The images in panel a were adapted from ref. 19, Springer Nature Ltd.
