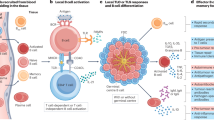
Abstract
Modern cell therapies are designed to harness natural biology to treat a range of complex diseases. The field of immunology has shown that B cells exhibit multiple unique features, including a natural propensity to interact with and regulate other immune cells, a high capacity to produce proteins, and a long cellular lifespan, which are being creatively applied in engineered B cell (eB cell) therapies. In recent years, advances in genome editing technologies and animal modeling have facilitated rapid progress in our ability to study eB cells and execute proof-of-concept studies, thus enabling the first clinical trials of eB cell therapies. In this review, we provide an overview of recent developments in eB cell therapies, including early clinical studies. We discuss challenges to clinical implementation, and promising directions for leveraging B cell biology in future applications for cancer and chronic disease.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ueda, N. et al. Immunotherapy perspectives in the new era of B-cell editing. Blood Adv. 5, 1770–1779 (2021).
Edelstein, J., Fritz, M. & Lai, S. K. Challenges and opportunities in gene editing of B cells. Biochem. Pharmacol. 206, 115285 (2022).
Jeske, A. M., Boucher, P., Curiel, D. & Voss, J. Vector strategies to actualize B cell-based gene therapies. J. Immunol. 207, 755–764 (2021).
Rogers, G. L. et al. Optimization of AAV6 transduction enhances site-specific genome editing of primary human lymphocytes. Mol. Ther. Methods Clin. Dev. 23, 198–209 (2021).
Page, A., Hubert, J., Fusil, F. & Cosset, F.-L. Exploiting B cell transfer for cancer therapy: engineered B cells to eradicate tumors. Int. J. Mol. Sci. 22, 9991 (2021).
Radbruch, A. et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat. Rev. Immunol. 6, 741–750 (2006).
Hibi, T. & Dosch, H. M. Limiting dilution analysis of the B cell compartment in human bone marrow. Eur. J. Immunol. 16, 139–145 (1986).
Amanna, I. J., Carlson, N. E. & Slifka, M. K. Duration of humoral immunity to common viral and vaccine antigens. N. Engl. J. Med. 357, 1903–1915 (2007).
Slifka, M. K., Antia, R., Whitmire, J. K. & Ahmed, R. Humoral immunity due to long-lived plasma cells. Immunity 8, 363–372 (1998).
Gorzelany, J. A. & de Souza, M. P. Protein replacement therapies for rare diseases: a breeze for regulatory approval? Sci. Transl. Med. 5, 178fs10 (2013).
Samelson-Jones, B. J. & George, L. A. Adeno-associated virus gene therapy for hemophilia. Annu. Rev. Med. 74, 231–247 (2023).
Srivastava, A. et al. Lentiviral gene therapy with CD34+ hematopoietic cells for hemophilia A. N. Engl. J. Med. 392, 450–457 (2025).
Tiede, A. Half-life extended factor VIII for the treatment of hemophilia A. J. Thromb. Haemost. 13, S176–S179 (2015).
Chen, H. H. et al. Enzyme replacement therapy for mucopolysaccharidoses; past, present, and future. J. Hum. Genet. 64, 1153–1171 (2019).
Chu, W. et al. Status and frontiers of Fabre disease. Orphanet J. Rare Dis. 20, 123 (2025).
Salabarria, S. M. et al. Advancements in AAV-mediated gene therapy for Pompe disease. J. Neuromuscul. Dis. 7, 15–31 (2020).
Placci, M., Giannotti, M. I. & Muro, S. Polymer-based drug delivery systems under investigation for enzyme replacement and other therapies of lysosomal storage disorders. Adv. Drug Deliv. Rev. 197, 114683 (2023).
Ozelo, M. C. et al. Valoctocogene roxaparvovec gene therapy for hemophilia A. N. Engl. J. Med. 386, 1013–1025 (2022).
Pipe, S. W. et al. Gene therapy with etranacogene dezaparvovec for hemophilia B. N. Engl. J. Med. 388, 706–718 (2023).
Mingozzi, F. & High, K. A. Overcoming the host immune response to adeno-associated virus gene delivery vectors: the race between clearance, tolerance, neutralization, and escape. Annu. Rev. Virol. 4, 511–534 (2017).
Cunningham, S. C., Dane, A. P., Spinoulas, A. & Alexander, I. E. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol. Ther. 16, 1081–1088 (2008).
Hill, T. F. et al. Human plasma cells engineered to secrete bispecifics drive effective in vivo leukemia killing. Mol. Ther. https://doi.org/10.1016/j.ymthe.2024.06.004 (2024).
Luo, B. et al. Engineering of α-PD-1 antibody-expressing long-lived plasma cells by CRISPR/Cas9-mediated targeted gene integration. Cell Death Dis. 11, 973 (2020).
Moffett, H. F. et al. B cells engineered to express pathogen-specific antibodies protect against infection. Sci. Immunol. 4, eaax0644 (2019).
Silacci, P., Mottet, A., Steimle, V., Reith, W. & Mach, B. Developmental extinction of major histocompatibility complex class II gene expression in plasmocytes is mediated by silencing of the transactivator gene CIITA. J. Exp. Med. 180, 1329–1336 (1994).
Duan, M. et al. Understanding heterogeneity of human bone marrow plasma cell maturation and survival pathways by single-cell analyses. Cell Rep. 42, 112682 (2023).
Young, D. J. et al. In vivo tracking of ex-vivo-generated 89Zr-oxine-labeled plasma cells by PET in a non-human primate model. Mol. Ther. https://doi.org/10.1016/j.ymthe.2024.12.042 (2024).
Hung, K. L. et al. Engineering protein-secreting plasma cells by homology-directed repair in primary human B cells. Mol. Ther. 26, 456–467 (2018).
David, M. et al. Production of therapeutic levels of human FIX-R338L by engineered B cells using GMP-compatible medium. Mol. Ther. Methods Clin. Dev. 31, 101111 (2023).
Liu, H. et al. A precision gene engineered B cell medicine producing sustained levels of active factor IX for hemophilia B therapy. Preprint at bioRxiv https://doi.org/10.1101/2025.04.06.647090 (2025).
Philippidis, A. Immusoft reports promising early data for lead candidate in MPS I. GEN—Genetic Engineering and Biotechnology News. www.genengnews.com/topics/drug-discovery/immusoft-reports-promising-early-data-for-lead-candidate-in-mps-i/ (2024).
Pipe, S. W. et al. Become-9: a phase 1/2 dose escalation and expansion study of be-101 for the treatment of adults with moderately severe or severe hemophilia B. Blood 144, 2593.1 (2024).
Rastogi, I. et al. Role of B cells as antigen presenting cells. Front. Immunol. 13, 954936 (2022).
Okada, T. et al. Antigen-engaged B cells undergo chemotaxis toward the T zone and form motile conjugates with helper T cells. PLoS Biol. 3, e150 (2005).
Song, W. & Craft, J. T follicular helper cell heterogeneity: time, space, and function. Immunol. Rev. 288, 85–96 (2019).
Elsner, R. A. & Shlomchik, M. J. Germinal center and extrafollicular B cell responses in vaccination, immunity, and autoimmunity. Immunity 53, 1136–1150 (2020).
Hartweger, H. et al. HIV-specific humoral immune responses by CRISPR/Cas9-edited B cells. J. Exp. Med. 216, 1301–1310 (2019).
Huang, D. et al. B cells expressing authentic naive human VRC01-class BCRs can be recruited to germinal centers and affinity mature in multiple independent mouse models. Proc. Natl Acad. Sci. USA 117, 22920–22931 (2020).
Huang, D. et al. Vaccine elicitation of HIV broadly neutralizing antibodies from engineered B cells. Nat. Commun. 11, 5850 (2020).
Nahmad, A. D. et al. Engineered B cells expressing an anti-HIV antibody enable memory retention, isotype switching and clonal expansion. Nat. Commun. 11, 5851 (2020).
Greiner, V. et al. CRISPR-mediated editing of the B cell receptor in primary human B cells. iScience 12, 369–378 (2019).
Voss, J. E. et al. Reprogramming the antigen specificity of B cells using genome-editing technologies. eLife 8, e42995 (2019).
Rogers, G. L. et al. Reprogramming human B cells with custom heavy-chain antibodies. Nat. Biomed. Eng. https://doi.org/10.1038/s41551-024-01240-4 (2024).
Yin, Y. et al. In vivo affinity maturation of mouse B cells reprogrammed to express human antibodies. Nat. Biomed. Eng. 8, 361–379 (2024).
Wennhold, K. et al. Using antigen-specific B cells to combine antibody and T cell-based cancer immunotherapy. Cancer Immunol. Res. 5, 730–743 (2017).
Lee-Chang, C. et al. Activation of 4-1BBL+ B cells with CD40 agonism and IFNγ elicits potent immunity against glioblastoma. J. Exp. Med. 218, e20200913 (2021).
Wang, S. et al. B cell-based therapy produces antibodies that inhibit glioblastoma growth. J. Clin. Invest. 134, e177384 (2024).
Winkler, J. et al. Adoptive transfer of donor B lymphocytes: a phase 1/2a study for patients after allogeneic stem cell transplantation. Blood Adv. 8, 2373–2383 (2024).
Winkler, J. et al. GMP-grade generation of B-lymphocytes for adoptive immunotherapy in patients after allogeneic stem cell transplantation. Blood 120, 4352 (2012).
Winkler, J. et al. Adoptive transfer of purified donor-B-lymphocytes after allogeneic stem cell transplantation: results from a phase I/IIa clinical trial. Blood 128, 502 (2016).
Fillatreau, S., Sweenie, C. H., McGeachy, M. J., Gray, D. & Anderton, S. M. B cells regulate autoimmunity by provision of IL-10. Nat. Immunol. 3, 944–950 (2002).
Shen, P. et al. IL-35-producing B cells are critical regulators of immunity during autoimmune and infectious diseases. Nature 507, 366–370 (2014).
Parekh, V. V. et al. B cells activated by lipopolysaccharide, but not by anti-Ig and anti-CD40 antibody, induce anergy in CD8+ T cells: role of TGF-β 1. J. Immunol. 170, 5897–5911 (2003).
Knippenberg, S. et al. Reduction in IL-10 producing B cells (Breg) in multiple sclerosis is accompanied by a reduced naive/memory Breg ratio during a relapse but not in remission. J. Neuroimmunol. 239, 80–86 (2011).
Blair, P. A. et al. CD19+CD24hiCD38hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity 32, 129–140 (2010).
Flores-Borja, F. et al. CD19+CD24hiCD38hi B cells maintain regulatory T cells while limiting TH1 and TH17 differentiation. Sci. Transl. Med. 5, 173ra23 (2013).
Rosser, E. C. & Mauri, C. Regulatory B cells: origin, phenotype, and function. Immunity 42, 607–612 (2015).
Shankar, S. et al. Ex vivo-expanded human CD19+TIM-1+ regulatory B cells suppress immune responses in vivo and are dependent upon the TIM-1/STAT3 axis. Nat. Commun. 13, 3121 (2022).
Lee, K. M. et al. Suppression of allograft rejection by regulatory B cells induced via TLR signaling. JCI Insight 7, e152213 (2022).
Bao, Y. et al. Ex vivo-generated human CD1c+ regulatory B cells by a chemically defined system suppress immune responses and alleviate graft-versus-host disease. Mol. Ther. 32, 4372–4382 (2024).
Zambidis, E. T., Kurup, A. & Scott, D. W. Genetically transferred central and peripheral immune tolerance via retroviral-mediated expression of immunogenic epitopes in hematopoietic progenitors or peripheral B lymphocytes. Mol. Med. 3, 212–224 (1997).
El-Amine, M. et al. Mechanisms of tolerance induction by a gene-transferred peptide-IgG fusion protein expressed in B lineage cells. J. Immunol. 165, 5631–5636 (2000).
Melo, M. E. F. et al. Gene transfer of Ig-fusion proteins into B cells prevents and treats autoimmune diseases. J. Immunol. 168, 4788–4795 (2002).
Song, L. et al. Retroviral delivery of GAD-IgG fusion construct induces tolerance and modulates diabetes: a role for CD4+ regulatory T cells and TGF-β? Gene Ther. 11, 1487–1496 (2004).
Lei, T. C. & Scott, D. W. Induction of tolerance to factor VIII inhibitors by gene therapy with immunodominant A2 and C2 domains presented by B cells as Ig fusion proteins. Blood 105, 4865–4870 (2005).
Wang, X. et al. Immune tolerance induction to factor IX through B cell gene transfer: TLR9 signaling delineates between tolerogenic and immunogenic B cells. Mol. Ther. 22, 1139–1150 (2014).
Ahangarani, R. R. et al. In vivo induction of type 1-like regulatory T cells using genetically modified B cells confers long-term IL-10-dependent antigen-specific unresponsiveness. J. Immunol. 183, 8232–8243 (2009).
Calderón-Gómez, E. et al. Reprogrammed quiescent B cells provide an effective cellular therapy against chronic experimental autoimmune encephalomyelitis. Eur. J. Immunol. 41, 1696–1708 (2011).
Chen, D. et al. Novel engineered B lymphocytes targeting islet-specific T cells inhibit the development of type 1 diabetes in non-obese diabetic Scid mice. Front. Immunol. 14, 1227133 (2023).
Pitner, R. A. et al. Blunting specific T-dependent antibody responses with engineered ‘decoy’ B cells. Mol. Ther. 32, 3453–3469 (2024).
Luo, X. M. et al. Engineering human hematopoietic stem/progenitor cells to produce a broadly neutralizing anti-HIV antibody after in vitro maturation to human B lymphocytes. Blood 113, 1422–1431 (2009).
Cheng, R. Y.-H. et al. Ex vivo engineered human plasma cells exhibit robust protein secretion and long-term engraftment in vivo. Nat. Commun. 13, 6110 (2022).
He, W. et al. Heavy-chain CDR3-engineered B cells facilitate in vivo evaluation of HIV-1 vaccine candidates. Immunity 56, 2408–2424.e6 (2023).
Serafini, M., Naldini, L. & Introna, M. Molecular evidence of inefficient transduction of proliferating human B lymphocytes by VSV-pseudotyped HIV-1-derived lentivectors. Virology 325, 413–424 (2004).
Janssens, W. et al. Efficiency of onco-retroviral and lentiviral gene transfer into primary mouse and human B-lymphocytes is pseudotype dependent. Hum. Gene Ther. 14, 263–276 (2003).
Frecha, C. et al. Efficient and stable transduction of resting B lymphocytes and primary chronic lymphocyte leukemia cells using measles virus gp displaying lentiviral vectors. Blood 114, 3173–3180 (2009).
Vamva, E. et al. A lentiviral vector B cell gene therapy platform for the delivery of the anti-HIV-1 eCD4-Ig-knob-in-hole-reversed immunoadhesin. Mol. Ther. Methods Clin. Dev. 28, 366–384 (2023).
Levy, C. et al. Baboon envelope pseudotyped lentiviral vectors efficiently transduce human B cells and allow active factor IX B cell secretion in vivo in NOD/SCIDγc−/− mice. J. Thromb. Haemost. 14, 2478–2492 (2016).
Bender, R. R. et al. Receptor-targeted Nipah virus glycoproteins improve cell-type selective gene delivery and reveal a preference for membrane-proximal cell attachment. PLoS Pathog. 12, e1005641 (2016).
Hamilton, J. R. et al. In vivo human T cell engineering with enveloped delivery vehicles. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-02085-z (2024).
Dobson, C. S. et al. Antigen identification and high-throughput interaction mapping by reprogramming viral entry. Nat. Methods 19, 449–460 (2022).
Yu, B. et al. Engineered cell entry links receptor biology with single-cell genomics. Cell 185, 4904–4920.e22 (2022).
Takano, K.-A. et al. Envelope protein-specific B cell receptors direct lentiviral vector tropism in vivo. Mol. Ther. 32, 1311–1327 (2024).
Ou, T. et al. Reprogramming of the heavy-chain CDR3 regions of a human antibody repertoire. Mol. Ther. 30, 184–197 (2022).
Johnson, M. J., Laoharawee, K., Lahr, W. S., Webber, B. R. & Moriarity, B. S. Engineering of primary human B cells with CRISPR/Cas9 targeted nuclease. Sci. Rep. 8, 12144 (2018).
Selvaraj, S. et al. High-efficiency transgene integration by homology-directed repair in human primary cells using DNA-PKcs inhibition. Nat. Biotechnol. 42, 731–744 (2024).
Sheridan, C. B cells as drug factories. Nat. Biotechnol. 42, 823–826 (2024).
Hackett, P. B. & Essner, J. Integration-site directed vector systems. US patent US7919583B2 (2005).
Laoharawee, K. et al. Genome engineering of primary human B cells using CRISPR/Cas9. J. Vis. Exp. https://doi.org/10.3791/61855 (2020).
Giguère, S. et al. Antibody production relies on the tRNA inosine wobble modification to meet biased codon demand. Science 383, 205–211 (2024).
Christie, S. M., Fijen, C., & Rothenberg, E. V(D)J recombination: recent insights in formation of the recombinase complex and recruitment of DNA repair machinery. Front. Cell Dev. Biol. 10, 886718 (2022).
Nahmad, A. D. et al. In vivo engineered B cells secrete high titers of broadly neutralizing anti-HIV antibodies in mice. Nat. Biotechnol. 40, 1241–1249 (2022).
Russell, D. M. et al. Peripheral deletion of self-reactive B cells. Nature 354, 308–311 (1991).
Abbott, R. K. et al. Precursor frequency and affinity determine B cell competitive fitness in germinal centers, tested with germline-targeting HIV vaccine immunogens. Immunity 48, 133–146.e6 (2018).
Dosenovic, P. et al. Anti–HIV-1 B cell responses are dependent on B cell precursor frequency and antigen-binding affinity. Proc. Natl Acad. Sci. USA 115, 4743–4748 (2018).
Tokatlian, T. et al. Enhancing humoral responses against HIV envelope trimers via nanoparticle delivery with stabilized synthetic liposomes. Sci. Rep. 8, 16527 (2018).
Wagar, L. E. et al. Modeling human adaptive immune responses with tonsil organoids. Nat. Med. 27, 125–135 (2021).
Pan, A. et al. In vivo affinity maturation of the CD4 domains of an HIV-1-entry inhibitor. Nat. Biomed. Eng. 8, 1715–1729 (2024).
Buerstedde, J.-M., Alinikula, J., Arakawa, H., McDonald, J. J. & Schatz, D. G. Targeting of somatic hypermutation by immunoglobulin enhancer and enhancer-like sequences. PLoS Biol. 12, e1001831 (2014).
O’Connor, B. P. et al. BCMA is essential for the survival of long-lived bone marrow plasma cells. J. Exp. Med. 199, 91–98 (2004).
Wallweber, H. J. A., Compaan, D. M., Starovasnik, M. A. & Hymowitz, S. G. The crystal structure of a proliferation-inducing ligand. April. J. Mol. Biol. 343, 283–290 (2004).
Lapidot, T. Mechanism of human stem cell migration and repopulation of NOD/SCID and B2mnull NOD/SCID mice. Ann. N. Y. Acad. Sci. 938, 83–95 (2001).
Hargreaves, D. C. et al. A coordinated change in chemokine responsiveness guides plasma cell movements. J. Exp. Med. 194, 45–56 (2001).
Chatterjee, S., Behnam Azad, B. & Nimmagadda, S. The intricate role of CXCR4 in cancer. Adv. Cancer Res. 124, 31–82 (2014).
Vonderheide, R. H., Tedder, T. F., Springer, T. A. & Staunton, D. E. Residues within a conserved amino acid motif of domains 1 and 4 of VCAM-1 are required for binding to VLA-4. J. Cell Biol. 125, 215–222 (1994).
Newham, P. et al. Α4 integrin binding interfaces on VCAM-1 and MAdCAM-1. J. Biol. Chem. 272, 19429–19440 (1997).
Benet, Z., Jing, Z. & Fooksman, D. R. Plasma cell dynamics in the bone marrow niche. Cell Rep. 34, 108733 (2021).
Nguyen, D. C. et al. Author correction: factors of the bone marrow microniche that support human plasma cell survival and immunoglobulin secretion. Nat. Commun. 10, 372 (2019).
Roldán, E., García-Pardo, A. & Brieva, J. A. VLA-4-fibronectin interaction is required for the terminal differentiation of human bone marrow cells capable of spontaneous and high rate immunoglobulin secretion. J. Exp. Med. 175, 1739–1747 (1992).
Fiorillo, M. T., Cabibbo, A., Iacopetti, P., Fattori, E. & Ciliberto, G. Analysis of human/mouse interleukin-6 hybrid proteins: both amino and carboxy termini of human interleukin-6 are required for in vitro receptor binding. Eur. J. Immunol. 22, 2609–2615 (1992).
Neuber, T. et al. Characterization and screening of IgG binding to the neonatal Fc receptor. MAbs 6, 928–942 (2014).
Ober, R. J., Radu, C. G., Ghetie, V. & Ward, E. S. Differences in promiscuity for antibody-FcRn interactions across species: implications for therapeutic antibodies. Int. Immunol. 13, 1551–1559 (2001).
Andersen, J. T., Daba, M. B., Berntzen, G., Michaelsen, T. E. & Sandlie, I. Cross-species binding analyses of mouse and human neonatal Fc receptor show dramatic differences in immunoglobulin G and albumin binding. J. Biol. Chem. 285, 4826–4836 (2010).
Li, F. et al. Mouse strains influence clearance and efficacy of antibody and antibody-drug conjugate via Fc-FcγR interaction. Mol. Cancer Ther. 18, 780–787 (2019).
Oldham, R. J. et al. FcγRII (CD32) modulates antibody clearance in NOD SCID mice leading to impaired antibody-mediated tumor cell deletion. J. Immunother. Cancer 8, e000619 (2020).
Yu, H. et al. A novel humanized mouse model with significant improvement of class-switched, antigen-specific antibody production. Blood 129, 959–969 (2017).
Li, Y. et al. A human immune system mouse model with robust lymph node development. Nat. Methods 15, 623–630 (2018).
Chupp, D. P. et al. A humanized mouse that mounts mature class-switched, hypermutated and neutralizing antibody responses. Nat. Immunol. https://doi.org/10.1038/s41590-024-01880-3 (2024).
Sun, K. & Liao, M. Z. Clinical pharmacology considerations on recombinant adeno-associated virus-based gene therapy. J. Clin. Pharmacol. 62, S79–S94 (2022).
Porter, D. L., Levine, B. L., Kalos, M., Bagg, A. & June, C. H. Chimeric antigen receptor-modified T cells in chronic lymphoid leukemia. N. Engl. J. Med. 365, 725–733 (2011).
Tsuchida, C. A. et al. Mitigation of chromosome loss in clinical CRISPR–Cas9-engineered T cells. Cell 186, 4567–4582.e20 (2023).
Lazar, N. H. et al. High-resolution genome-wide mapping of chromosome-arm-scale truncations induced by CRISPR-Cas9 editing. Nat. Genet. 56, 1482–1493 (2024).
Nahmad, A. D. et al. Frequent aneuploidy in primary human T cells after CRISPR–Cas9 cleavage. Nat. Biotechnol. 40, 1807–1813 (2022).
Zhang, T.-T. et al. BCR signaling is required for posttransplant lymphoproliferative disease in immunodeficient mice receiving human B cells. Sci. Transl. Med. 16, eadh8846 (2024).
Stockfelt, M., Teng, Y. K. O. & Vital, E. M. Opportunities and limitations of B cell depletion approaches in SLE. Nat. Rev. Rheumatol. 21, 111–126 (2025).
Gargett, T. & Brown, M. P. The inducible caspase-9 suicide gene system as a “safety switch†to limit on-target, off-tumor toxicities of chimeric antigen receptor T cells. Front. Pharmacol. 5, 235 (2014).
Dunkelberger, J. R. & Song, W.-C. Complement and its role in innate and adaptive immune responses. Cell Res. 20, 34–50 (2010).
Ma, A. D. & Carrizosa, D. Acquired factor VIII inhibitors: pathophysiology and treatment. Hematol. Am. Soc. Hematol. Educ. Program 2006, 432–437 (2006).
Jawa, V. et al. T-cell dependent immunogenicity of protein therapeutics pre-clinical assessment and mitigation-updated consensus and review 2020. Front. Immunol. 11, 1301 (2020).
Sabatino, D. E. et al. Efficacy and safety of long-term prophylaxis in severe hemophilia A dogs following liver gene therapy using AAV vectors. Mol. Ther. 19, 442–449 (2011).
Annoni, A. et al. Liver gene therapy by lentiviral vectors reverses anti-factor IX pre-existing immunity in haemophilic mice. EMBO Mol. Med. 5, 1684–1697 (2013).
Chand, D. et al. Hepatotoxicity following administration of onasemnogene abeparvovec (AVXS-101) for the treatment of spinal muscular atrophy. J. Hepatol. 74, 560–566 (2021).
Manno, C. S. et al. Successful transduction of liver in hemophilia by AAV-Factor IX and limitations imposed by the host immune response. Nat. Med. 12, 342–347 (2006).
Gallo-Penn, A. M. et al. Systemic delivery of an adenoviral vector encoding canine factor VIII results in short-term phenotypic correction, inhibitor development, and biphasic liver toxicity in hemophilia A dogs. Blood 97, 107–113 (2001).
Grauwet, K. et al. Stealth transgenes enable CAR-T cells to evade host immune responses. J. Immunother. Cancer 12, e008417 (2024).
Wang, B. et al. Generation of hypoimmunogenic T cells from genetically engineered allogeneic human induced pluripotent stem cells. Nat. Biomed. Eng. 5, 429–440 (2021).
Hu, X. et al. Hypoimmune induced pluripotent stem cells survive long term in fully immunocompetent, allogeneic rhesus macaques. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01784-x (2023).
Gupta, P., Alheib, O. & Shin, J.-W. Towards single cell encapsulation for precision biology and medicine. Adv. Drug Deliv. Rev. 201, 115010 (2023).
Kershaw, M. H. et al. A phase I study on adoptive immunotherapy using gene-modified T cells for ovarian cancer. Clin. Cancer Res. 12, 6106–6115 (2006).
Hartweger, H. et al. Gene editing of primary rhesus macaque B cells. J. Vis. Exp. https://doi.org/10.3791/64858 (2023).
Vamva, E. et al. An optimized measles virus glycoprotein-pseudotyped lentiviral vector production system to promote efficient transduction of human primary B cells. STAR Protoc. 3, 101228 (2022).
Yu-Hong Cheng, R. et al. Generation, expansion, gene delivery, and single-cell profiling in rhesus macaque plasma B cells. Cell Rep. Methods 4, 100878 (2024).
Ishikawa, M. et al. Bone marrow plasma cells require P2RX4 to sense extracellular ATP. Nature 626, 1102–1107 (2024).
Van Dam, M. et al. Structure–function analysis of interleukin-6 utilizing human/murine chimeric molecules. Involvement of two separate domains in receptor binding. J. Biol. Chem. 268, 15285–15290 (1993).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
R.G.J. and D.J.R. have an equity ownership position in and serve on the scientific advisory board of Be Biopharma. R.A.P., R.G.J. and D.J.R. are credited as inventors of patents filed by Seattle Children’s Research Institute with the United States Patent and Trademark Office regarding technologies described in this article (US20240287452A1, WO2025064484A1, US20230272431A1 and WO2023225447A1).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Table 1 and Supplementary Fig. 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Trivedi, N., Pitner, R.A., Rawlings, D.J. et al. Engineering B cells to treat and study human disease. Nat Biotechnol 43, 1431–1444 (2025). https://doi.org/10.1038/s41587-025-02757-y
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41587-025-02757-y