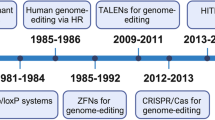
Abstract
Genome editing has revolutionized the treatment of genetic diseases, yet the difficulty of tissue-specific delivery currently limits applications of editing technology. In this Review, we discuss preclinical and clinical advances in delivering genome editors with both established and emerging delivery mechanisms. Targeted delivery promises to considerably expand the therapeutic applicability of genome editing, moving closer to the ideal of a precise ‘magic bullet’ that safely and effectively treats diverse genetic disorders.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Levesque, S. & Bauer, D. E. CRISPR-based therapeutic genome editing for inherited blood disorders. Nat. Rev. Drug Discov. https://doi.org/10.1038/s41573-025-01236-y (2025).
Raguram, A., Banskota, S. & Liu, D. R. Therapeutic in vivo delivery of gene editing agents. Cell 185, 2806–2827 (2022).
Tsuchida, C. A., Wasko, K. M., Hamilton, J. R. & Doudna, J. A. Targeted nonviral delivery of genome editors in vivo. Proc. Natl Acad. Sci. USA 121, e2307796121 (2024).
Stigzelius, V., Cavallo, A. L., Chandode, R. K. & Nitsch, R. Peeling back the layers of immunogenicity in Cas9-based genomic medicine. Mol. Ther. 33, 4714–4730 (2025).
Porteus, M. Genome editing: a new approach to human therapeutics. Annu. Rev. Pharmacol. Toxicol. 56, 163–190 (2014).
Kumar, M., Kulkarni, P., Liu, S., Chemuturi, N. & Shah, D. K. Nanoparticle biodistribution coefficients: a quantitative approach for understanding the tissue distribution of nanoparticles. Adv. Drug Deliv. Rev. 194, 114708 (2023).
Gillmore, J. D. et al. CRISPR–Cas9 in vivo gene editing for transthyretin amyloidosis. N. Engl. J. Med. 385, 493–502 (2021).
Musunuru, K. et al. Patient-specific in vivo gene editing to treat a rare genetic disease. N. Engl. J. Med. 392, 2235–2243 (2025).
Horie, T. & Ono, K. VERVE-101: a promising CRISPR-based gene editing therapy that reduces LDL-C and PCSK9 levels in HeFH patients. Eur. Heart J. Cardiovasc. Pharmacother. 10, 89–90 (2023).
Cohn, D. M. et al. CRISPR-based therapy for hereditary angioedema. N. Engl. J. Med. 392, 458–467 (2025).
Lee, R. et al. An investigational in vivo base editing medicine targeting ANGPTL3, VERVE-201, achieves precise and durable liver editing in nonclinical studies. Atherosclerosis 395, 118496 (2024).
Beam Therapeutics. A phase 1/2 dose-exploration and dose-expansion study to evaluate the safety and efficacy of BEAM-302 in adult patients with α-1 antitrypsin deficiency (AATD)-associated lung disease and/or liver disease. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06389877 (2024).
Morrow, P. K. et al. Abstract 17013: CTX320: an investigational in vivo CRISPR-based therapy efficiently and durably reduces lipoprotein (a) levels in non-human primates after a single dose. Circulation 148, A17013 (2023).
HuidaGene Therapeutics. An investigator-initiated clinical study evaluating the CRISPR–hfCas12Max gene editing therapy in the treatment of Duchenne muscular dystrophy (DMD). ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06594094 (2024).
Beam Therapeutics. A phase 1/2, dose-exploration study to evaluate the safety and efficacy of BEAM-301 in patients with glycogen storage disease type Ia (GSDIa) homozygous or compound heterozygous for the G6PC1 c.247C>T (p.R83C) variant. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06735755 (2024).
Arbor Biotechnologies. A phase 1/2 dose escalation study to evaluate the safety, tolerability, pharmacokinetics, pharmacodynamics and preliminary efficacy of ABO-101 in participants with primary hyperoxaluria type 1 (PH1). ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06839235 (2025).
Tune Therapeutics. Phase 1b multicenter, open-label study to assess the safety, tolerability, pharmacokinetics, and pharmacodynamics of Tune-401 in participants with chronic hepatitis B infection. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06671093 (2024).
Burdo, T. H. et al. Preclinical safety and biodistribution of CRISPR targeting SIV in non-human primates. Gene Ther. 31, 224–233 (2023).
Excision BioTherapeutics. A phase 1/2a, sequential cohort, single ascending dose study of the safety, tolerability, biodistribution, and pharmacodynamics of EBT 101 in aviremic HIV-1 infected adults on stable antiretroviral therapy. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT05144386 (2021).
Epicrispr Biotechnologies. A phase 1/2 open-label dose-escalation study to evaluate the safety, tolerability, and biological activity of EPI-321, an AAVrh74-delivered epigenetic editing therapy in adult FSHD patients. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06907875 (2025).
Streilein, J. W. Ocular immune privilege: therapeutic opportunities from an experiment of nature. Nat. Rev. Immunol. 3, 879–889 (2003).
Nakao, S., Hafezi-Moghadam, A. & Ishibashi, T. Lymphatics and lymphangiogenesis in the eye. J. Ophthalmol. 2012, 783163 (2012).
Toral, M. A. et al. Investigation of Cas9 antibodies in the human eye. Nat. Commun. 13, 1053 (2022).
Pierce, E. A. et al. Gene editing for CEP290-associated retinal degeneration. N. Engl. J. Med. 390, 1972–1984 (2024).
Zhao, Q., Wei, L. & Chen, Y. From bench to bedside: developing CRISPR/Cas-based therapy for ocular diseases. Pharmacol. Res. 213, 107638 (2025).
Muller, A. et al. High-efficiency base editing in the retina in primates and human tissues. Nat. Med. 31, 490–501 (2025).
Luk, A. et al. World’s first CRISPR/RNA-targeting therapy (HG202) for patients with neovascular age-related macular degeneration. Invest. Ophthalmol. Vis. Sci. 65, 4357 (2024).
Wei, A. et al. In vivo CRISPR gene editing in patients with herpetic stromal keratitis. Mol. Ther. 31, 3163–3175 (2023).
Jain, A. et al. CRISPR–Cas9–based treatment of myocilin-associated glaucoma. Proc. Natl Acad. Sci. USA 114, 11199–11204 (2017).
Gencay, Y. E. et al. Engineered phage with antibacterial CRISPR–Cas selectively reduce E. coli burden in mice. Nat. Biotechnol. 42, 265–274 (2024).
SNIPR Biome. A phase 1, randomized, double-blind, first-in-human, dose escalation study investigating the safety, recovery, and pharmacodynamics of multiple oral administrations of SNIPR001 in healthy subjects. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT05277350 (2022).
SNIPR Biome. SNIPR Biome Reports Positive Clinical Interim Results for Groundbreaking, First-in-Human, CRISPR-Based Microbial Gene Therapy https://static1.squarespace.com/static/5bacc67990f9041ab0d5b0c1/t/6476ee0c6181141d414b9ec3/1685515789399/230529+SNIPR+Phase+1+Data+Release.pdf (2023).
Xue, Y. et al. RNA base editing therapy cures hearing loss induced by OTOF gene mutation. Mol. Ther. 31, 3520–3530 (2023).
HuidaGene Therapeutics. An open-label, multiple-cohort, dose-finding, investigator-initiated trial to evaluate the safety, tolerability, and efficacy of HG205 RNA base-editing therapy in subjects with OTOF-p.Q829X mutation-associated hearing loss. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06025032 (2023).
Yang, D. et al. An RNA editing strategy rescues gene duplication in a mouse model of MECP2 duplication syndrome and nonhuman primates. Nat. Neurosci. 28, 72–83 (2025).
HuidaGene Therapeutics. An open-label, multiple-dose clinical study to evaluating the safety, tolerability and preliminary efficacy of a single intracerebroventricular injection of HG204 for the treatment of MECP2 duplication syndrome. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06615206 (2024).
Lenneman, B. R., Fernbach, J., Loessner, M. J., Lu, T. K. & Kilcher, S. Enhancing phage therapy through synthetic biology and genome engineering. Curr. Opin. Biotechnol. 68, 151–159 (2021).
Kim, P. et al. Safety, pharmacokinetics, and pharmacodynamics of LBP-EC01, a CRISPR–Cas3-enhanced bacteriophage cocktail, in uncomplicated urinary tract infections due to Escherichia coli (ELIMINATE): the randomised, open-label, first part of a two-part phase 2 trial. Lancet Infect. Dis. 24, 1319–1332 (2024).
Amoasii, L. et al. Single-cut genome editing restores dystrophin expression in a new mouse model of muscular dystrophy. Sci. Transl. Med. 9, eaan8081 (2017).
Ho, T.-C. et al. Scaffold-mediated CRISPR–Cas9 delivery system for acute myeloid leukemia therapy. Sci. Adv. 7, eabg3217 (2021).
Liang, S.-Q. et al. AAV5 delivery of CRISPR–Cas9 supports effective genome editing in mouse lung airway. Mol. Ther. 30, 238–243 (2022).
Rosenblum, D. et al. CRISPR–Cas9 genome editing using targeted lipid nanoparticles for cancer therapy. Sci. Adv. 6, eabc9450 (2020).
Stahl, E. C. et al. Genome editing in the mouse brain with minimally immunogenic Cas9 RNPs. Mol. Ther. 31, 2422–2438 (2023).
Kasiewicz, L. N. et al. GalNAc-lipid nanoparticles enable non-LDLR dependent hepatic delivery of a CRISPR base editing therapy. Nat. Commun. 14, 2776 (2023).
Lee, R. et al. An investigational in vivo base editing medicine targeting ANGPTL3, VERVE-201, achieves potent and LDLR-independent liver editing in mouse models. Eur. Heart J. 44, ehad655.2521 (2023).
Verve Therapeutics. A phase 1b single ascending dose study to evaluate the safety of VERVE-201 in patients with refractory hyperlipidemia. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06451770 (2024).
Pupo, A. et al. AAV vectors: the Rubik’s cube of human gene therapy. Mol. Ther. 30, 3515–3541 (2022).
Gao, G.-P. et al. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl Acad. Sci. USA 99, 11854–11859 (2002).
Strebinger, D. et al. Cell type-specific delivery by modular envelope design. Nat. Commun. 14, 5141 (2023).
Hamilton, J. R. et al. In vivo human T cell engineering with enveloped delivery vehicles. Nat. Biotechnol. 42, 1684–1692 (2024).
Hamilton, J. R. et al. Targeted delivery of CRISPR–Cas9 and transgenes enables complex immune cell engineering. Cell Rep. 35, 109207 (2021).
Ngo, W. et al. Mechanism-guided engineering of a minimal biological particle for genome editing. Proc. Natl Acad. Sci. USA 122, e2413519121 (2025).
Karp, H. et al. Packaged delivery of CRISPR–Cas9 ribonucleoproteins accelerates genome editing. Nucleic Acids Res. 53, gkaf105 (2025).
Breda, L. et al. In vivo hematopoietic stem cell modification by mRNA delivery. Science 381, 436–443 (2023).
Palanki, R. et al. In utero delivery of targeted ionizable lipid nanoparticles facilitates in vivo gene editing of hematopoietic stem cells. Proc. Natl Acad. Sci. USA 121, e2400783121 (2024).
Geczy, R. et al. Lipid nanoparticle-mediated gene editing of human primary T cells and off-target analysis of the CRISPR–Cas9 indels. Blood 142, 6833 (2023).
Dilliard, S. A., Cheng, Q. & Siegwart, D. J. On the mechanism of tissue-specific mRNA delivery by selective organ targeting nanoparticles. Proc. Natl Acad. Sci. USA 118, e2109256118 (2021).
Cheng, Q. et al. Selective organ targeting (SORT) nanoparticles for tissue specific mRNA delivery and CRISPR/Cas gene editing. Nat. Nanotechnol. 15, 313–320 (2020).
Chen, K. et al. Lung and liver editing by lipid nanoparticle delivery of a stable CRISPR–Cas9 ribonucleoprotein. Nat. Biotechnol. 43, 1445–1457 (2025).
Kimura, S. & Harashima, H. On the mechanism of tissue-selective gene delivery by lipid nanoparticles. J. Control. Release 362, 797–811 (2023).
Tabebordbar, M. et al. Directed evolution of a family of AAV capsid variants enabling potent muscle-directed gene delivery across species. Cell 184, 4919–4938 (2021).
Huang, Q. et al. An AAV capsid reprogrammed to bind human transferrin receptor mediates brain-wide gene delivery. Science 384, 1220–1227 (2024).
Neumann, E. N. et al. Brainwide silencing of prion protein by AAV-mediated delivery of an engineered compact epigenetic editor. Science 384, ado7082 (2024).
Kumar, S. R. et al. Multiplexed Cre-dependent selection yields systemic AAVs for targeting distinct brain cell types. Nat. Methods 17, 541–550 (2020).
Kim, H. et al. Lipid nanoparticle-mediated mRNA delivery to CD34+ cells in rhesus monkeys. Nat. Biotechnol. 43, 1813–1820 (2024).
Dahlman, J. E. et al. Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. Proc. Natl Acad. Sci. USA 114, 2060–2065 (2017).
Ngo, W. et al. Why nanoparticles prefer liver macrophage cell uptake in vivo. Adv. Drug Deliv. Rev. 185, 114238 (2022).
Glaumann, H., Fredzell, J., Jubner, A. & Ericsson, J. L. E. Uptake and degradation of glycogen by Kupffer cells. Exp. Mol. Pathol. 31, 70–80 (1979).
Seo, J. W. et al. Multimodal imaging of capsid and cargo reveals differential brain targeting and liver detargeting of systemically-administered AAVs. Biomaterials 288, 121701 (2022).
l’Hortet, A. C. et al. In MDA Clinical & Scientific Conference 206 https://www.mdaconference.org/abstract-library/epi-321-a-potential-cure-for-fshd/ (Muscular Dystrophy Association, 2023).
Amoasii, L. et al. Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86–91 (2018).
Vaessen, S. F. C. et al. AAV gene therapy as a means to increase apolipoprotein (Apo) A-I and high-density lipoprotein-cholesterol levels: correction of murine ApoA-I deficiency. J. Gene Med. 11, 697–707 (2009).
Prasad, K.-M. R., Xu, Y., Yang, Z., Acton, S. T. & French, B. A. Robust cardiomyocyte-specific gene expression following systemic injection of AAV: in vivo gene delivery follows a Poisson distribution. Gene Ther. 18, 43–52 (2011).
Radhiyanti, P. T., Konno, A., Matsuzaki, Y. & Hirai, H. Comparative study of neuron-specific promoters in mouse brain transduced by intravenously administered AAV-PHP.eB. Neurosci. Lett. 756, 135956 (2021).
Yang, L. et al. MicroRNA-122-mediated liver detargeting enhances the tissue specificity of cardiac genome editing. Circulation 149, 1778–1781 (2024).
Hoffmann, M. D. et al. Cell-specific CRISPR–Cas9 activation by microRNA-dependent expression of anti-CRISPR proteins. Nucleic Acids Res. 47, e75 (2019).
Hirosawa, M., Fujita, Y. & Saito, H. Cell-type-specific CRISPR activation with microRNA-responsive AcrllA4 switch. ACS Synth. Biol. 8, 1575–1582 (2019).
Lee, J. et al. Tissue-restricted genome editing in vivo specified by microRNA-repressible anti-CRISPR proteins. RNA 25, 1421–1431 (2019).
Wang, X.-W. et al. A microRNA-inducible CRISPR–Cas9 platform serves as a microRNA sensor and cell-type-specific genome regulation tool. Nat. Cell Biol. 21, 522–530 (2019).
Garcia-Guerra, A. et al. Tissue-specific modulation of CRISPR activity by miRNA-sensing guide RNAs. Nucleic Acids Res. 53, gkaf016 (2025).
Galizi, R. & Jaramillo, A. Engineering CRISPR guide RNA riboswitches for in vivo applications. Curr. Opin. Biotechnol. 55, 103–113 (2019).
Kaseniit, K. E. et al. Modular, programmable RNA sensing using ADAR editing in living cells. Nat. Biotechnol. 41, 482–487 (2023).
Jiang, K. et al. Programmable eukaryotic protein synthesis with RNA sensors by harnessing ADAR. Nat. Biotechnol. 41, 698–707 (2023).
Qian, Y. et al. Programmable RNA sensing for cell monitoring and manipulation. Nature 610, 713–721 (2022).
Powell, S. K., Rivera-Soto, R. & Gray, S. J. Viral expression cassette elements to enhance transgene target specificity and expression in gene therapy. Discov. Med. 19, 49–57 (2015).
Mancuso, P. et al. CRISPR based editing of SIV proviral DNA in ART treated non-human primates. Nat. Commun. 11, 6065 (2020).
Cohrt, K. O. Excision’s EBT-101 demonstrates safety in clinical trial but does not cure HIV. CRISPR Medicine News https://crisprmedicinenews.com/news/excisions-ebt-101-demonstrates-safety-in-clinical-trial-but-does-not-cure-hiv/ (2024).
Tan, I.-L. et al. Targeting the non-coding genome and temozolomide signature enables CRISPR-mediated glioma oncolysis. Cell Rep. 42, 113339 (2023).
An, Y. et al. Design of hypoxia responsive CRISPR–Cas9 for target gene regulation. Sci. Rep. 13, 16763 (2023).
Chen, X., Chen, Y., Xin, H., Wan, T. & Ping, Y. Near-infrared optogenetic engineering of photothermal nanoCRISPR for programmable genome editing. Proc. Natl Acad. Sci. USA 117, 2395–2405 (2020).
Yin, H. et al. Ultrasound-controlled CRISPR/Cas9 system augments sonodynamic therapy of hepatocellular carcinoma. ACS Cent. Sci. 7, 2049–2062 (2021).
Liu, Y. et al. Very fast CRISPR on demand. Science 368, 1265–1269 (2020).
Pacesa, M. et al. Structural basis for Cas9 off-target activity. Cell 185, 4067–4081 (2022).
Greig, J. A. et al. Integrated vector genomes may contribute to long-term expression in primate liver after AAV administration. Nat. Biotechnol. 42, 1232–1242 (2024).
iECURE. A phase 1/2/3 first-in-human, open-label, dose-escalation study to evaluate the safety and efficacy of a single intravenous (IV) administration of ECUR-506 in males less than 9 months of age with genetically confirmed neonatal onset ornithine transcarbamylase (OTC) deficiency. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06255782 (2023).
Regeneron Pharmaceuticals. A two-part open-label study of REGV131-LNP1265, a CRISPR/Cas9 based coagulation factor IX gene insertion therapy in participants with hemophilia B. ClinicalTrials.gov https://clinicaltrials.gov/study/NCT06379789 (2024).
Jeune, V. L., Joergensen, J. A., Hajjar, R. J. & Weber, T. Pre-existing anti-adeno-associated virus antibodies as a challenge in AAV gene therapy. Hum. Gene Ther. Methods 24, 59–67 (2013).
Duan, D. Lethal immunotoxicity in high-dose systemic AAV therapy. Mol. Ther. 31, 3123–3126 (2023).
Lee, Y., Jeong, M., Park, J., Jung, H. & Lee, H. Immunogenicity of lipid nanoparticles and its impact on the efficacy of mRNA vaccines and therapeutics. Exp. Mol. Med. 55, 2085–2096 (2023).
Vargas, J. E. et al. Retroviral vectors and transposons for stable gene therapy: advances, current challenges and perspectives. J. Transl. Med. 14, 288 (2016).
Wignakumar, T. & Fairchild, P. J. Evasion of pre-existing immunity to Cas9: a prerequisite for successful genome editing in vivo? Curr. Transplant. Rep. 6, 127–133 (2019).
Kishimoto, T. K. & Samulski, R. J. Addressing high dose AAV toxicity — ‘one and done’ or ‘slower and lower’? Expert Opin. Biol. Ther. 22, 1067–1071 (2022).
Cullis, P. R. & Hope, M. J. Lipid nanoparticle systems for enabling gene therapies. Mol. Ther. 25, 1467–1475 (2017).
Cullis, P. R. & Felgner, P. L. The 60-year evolution of lipid nanoparticles for nucleic acid delivery. Nat. Rev. Drug Discov. 23, 709–722 (2024).
Carbonaro-Sarracino, D. A. et al. Dosing and re-administration of intravenous lentiviral vector for liver-directed gene transfer in young rhesus monkeys and ADA-deficient mice. Mol. Ther. Methods Clin. Dev. 24, S302–S303 (2016).
Chen, K. et al. Engineering self-deliverable ribonucleoproteins for genome editing in the brain. Nat. Commun. 15, 1727 (2024).
Staahl, B. T. et al. Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat. Biotechnol. 35, 431–434 (2017).
Chew, W. L. Immunity to CRISPR Cas9 and Cas12a therapeutics. Wiley Interdiscip. Rev. Syst. Biol. Med. https://doi.org/10.1002/wsbm.1408 (2018).
Andari, J. E. & Grimm, D. Production, processing, and characterization of synthetic AAV gene therapy vectors. Biotechnol. J. 16, e2000025 (2021).
Jiang, Z. & Dalby, P. A. Challenges in scaling up AAV-based gene therapy manufacturing. Trends Biotechnol. 41, 1268–1281 (2023).
De, A. & Ko, Y. T. Why mRNA-ionizable LNPs formulations are so short-lived: causes and way-out. Expert Opin. Drug Deliv. 20, 175–187 (2023).
Kim, B. et al. Optimization of storage conditions for lipid nanoparticle-formulated self-replicating RNA vaccines. J. Control. Release 353, 241–253 (2023).
Mangeot, P. E. et al. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9–sgRNA ribonucleoproteins. Nat. Commun. 10, 45 (2019).
Merten, O.-W., Hebben, M. & Bovolenta, C. Production of lentiviral vectors. Mol. Ther. Methods Clin. Dev. 3, 16017 (2016).
Binder, G. K. & Chen, C.-C. The very stable lentiviral vector. Mol. Ther. Methods Clin. Dev. 32, 101223 (2024).
Berry, G. E. & Asokan, A. Cellular transduction mechanisms of adeno-associated viral vectors. Curr. Opin. Virol. 21, 54–60 (2016).
Patel, M. N. et al. Safer non-viral DNA delivery using lipid nanoparticles loaded with endogenous anti-inflammatory lipids. Nat. Biotechnol. https://doi.org/10.1038/s41587-025-02556-5 (2025).
Banskota, S. et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 185, 250–265 (2022).
An, M. et al. Engineered virus-like particles for transient delivery of prime editor ribonucleoprotein complexes in vivo. Nat. Biotechnol. 42, 1526–1537 (2024).
Lyu, P., Javidi-Parsijani, P., Atala, A. & Lu, B. Delivering Cas9/sgRNA ribonucleoprotein (RNP) by lentiviral capsid-based bionanoparticles for efficient ‘hit-and-run’ genome editing. Nucleic Acids Res. 47, e99 (2019).
Indikova, I. & Indik, S. Highly efficient ‘hit-and-run’ genome editing with unconcentrated lentivectors carrying Vpr.Prot.Cas9 protein produced from RRE-containing transcripts. Nucleic Acids Res. 48, 8178–8187 (2020).
Gao, G., Vandenberghe, L. H. & Wilson, J. M. New recombinant serotypes of AAV vectors. Curr. Gene Ther. 5, 285–297 (2005).
Pham, Q. et al. A facile chemical strategy to synthesize precise AAV-protein conjugates for targeted gene delivery. Mol. Ther. Oncol. 33, 201040 (2025).
Domenger, C. & Grimm, D. Next-generation AAV vectors—do not judge a virus (only) by its cover. Hum. Mol. Genet. 28, R3–R14 (2019).
Billingsley, M. M. et al. In vivo mRNA CAR T cell engineering via targeted ionizable lipid nanoparticles with extrahepatic tropism. Small 20, e2304378 (2024).
Rurik, J. G. et al. CAR T cells produced in vivo to treat cardiac injury. Science 375, 91–96 (2022).
Veiga, N. et al. Cell specific delivery of modified mRNA expressing therapeutic proteins to leukocytes. Nat. Commun. 9, 4493 (2018).
Dobson, C. S. et al. Antigen identification and high-throughput interaction mapping by reprogramming viral entry. Nat. Methods 19, 449–460 (2022).
Girard-Gagnepain, A. et al. Baboon envelope pseudotyped LVs outperform VSV-G-LVs for gene transfer into early-cytokine-stimulated and resting HSCs. Blood 124, 1221–1231 (2014).
Seydel, C. Spotlight Therapeutics: making CRISPR deliver in vivo. Nat. Biotechnol. https://doi.org/10.1038/d41587-021-00011-9 (2021).
Acknowledgements
We thank all members of the Doudna laboratory for their thoughtful input on the manuscript, especially K. Chen, J. Zeng and Z.W. Xue. The manuscript was funded by the following sources: Gladstone Institutes (J.A.D.), the HHMI (J.A.D.), National Heart, Lung, and Blood Institute grant 1R21HL173710-01 (J.A.D.) and Lawrence Livermore National Labs PROTECT grant, DE-AC52-07NA27344 (J.A.D.). J.A.D. also receives support from NIH/NIAID (U54AI170792, U19AI135990, UH3AI150552 and U01AI142817), NIH/NINDS (U19NS132303), NSF (2334028), DOE (DE-AC02-05CH11231, 2553571 and B656358), Apple Tree Partners (24180), UCB-Hampton University Summer Program, Mr. Li Ka Shing, Koret-Berkeley-TAU, Emerson Collective and the Innovative Genomics Institute (IGI). J.L.Y.W. was also funded by the Natural Sciences and Engineering Research Council of Canada Postdoctoral Fellowship. We also acknowledge financial support from the James B. Pendleton Charitable Trust.
Author information
Authors and Affiliations
Contributions
Conceptualization: W.N., K.M.W. and J.A.D. Funding acquisition: J.A.D. Project administration: W.N. and J.A.D. Writing and editing: W.N., K.M.W., J.L.Y.W. and J.A.D.
Corresponding author
Ethics declarations
Competing interests
The Regents of the University of California have patents issued and/or pending for CRISPR technologies (on which J.A.D. is an inventor) and delivery technologies (on which J.A.D. and W.N. are co-inventors). J.A.D. is a cofounder of Azalea Therapeutics, Caribou Biosciences, Editas Medicine, Evercrisp, Scribe Therapeutics and Mammoth Biosciences. J.A.D. is a scientific advisory board member at Isomorphic Labs, BEVC Management, Evercrisp, Caribou Biosciences, Scribe Therapeutics, Mammoth Biosciences, The Column Group and Inari. She is also an advisor for Aditum Bio. J.A.D. is chief science advisor to Sixth Street, is a director at Johnson & Johnson, Altos and Tempus, and has a research project sponsored by Apple Tree Partners. All other authors declare no competing interests.
Peer review
Peer review information
Nature Biotechnology thanks Dan Peer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ngo, W., Wu, J.L.Y., Wasko, K.M. et al. Targeted delivery of genome editors in vivo. Nat Biotechnol 44, 49–59 (2026). https://doi.org/10.1038/s41587-025-02945-w
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41587-025-02945-w