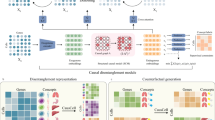
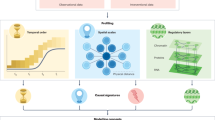
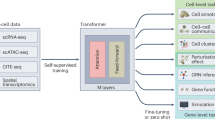
Abstract
Advances in single-cell '-omics' allow unprecedented insights into the transcriptional profiles of individual cells and, when combined with large-scale perturbation screens, enable measuring of the effect of targeted perturbations on the whole transcriptome. These advances provide an opportunity to better understand the causative role of genes in complex biological processes. In this Perspective, we delineate the application of causal machine learning to single-cell genomics and its associated challenges. We first present the causal model that is most commonly applied to single-cell biology and then identify and discuss potential approaches to three open problems: the lack of generalization of models to novel experimental conditions, the complexity of interpreting learned models, and the difficulty of learning cell dynamics.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
McInnes, L., Healy, J., Saul, N. & Großberger, L. UMAP: Uniform Manifold Approximation and Projection. J. Open Source Softw. 3, 861 (2018).
van der Maaten, L. & Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 9, 2579–2605 (2008).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Lange, M. et al. CellRank for directed single-cell fate mapping. Nat. Methods 19, 159–170 (2022).
Hetzel, L. et al. Predicting cellular responses to novel drug perturbations at a single-cell resolution. In Proceedings of the 36th International Conference on Neural Information Processing Systems 26711–26722 (Curran Associates, 2022).
Liu, J. et al. Towards out-of-distribution generalization: a survey. Preprint at https://arxiv.org/abs/2108.13624 (2021).
Sekhon, J. The Neyman–Rubin model of causal inference and estimation via matching methods. In The Oxford Handbook of Political Methodology (eds Box-Steffensmeier, J. M. et al.) Ch. 11 (Oxford Academic, 2008).
Imbens, G. W. & Rubin, D. B. Causal Inference in Statistics, Social, and Biomedical Sciences (Cambridge University Press, 2015).
Segal, E. et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 34, 166–176 (2003).
Qiao, L., Khalilimeybodi, A., Linden-Santangeli, N. J. & Rangamani, P. The evolution of systems biology and systems medicine: from mechanistic models to uncertainty quantification. Annu. Rev. Biomed. Eng. https://doi.org/10.1146/annurev-bioeng-102723-065309 (2025).
Wen, Y. et al. Applying causal discovery to single-cell analyses using CausalCell. eLife 12, e81464 (2023).
Belyaeva, A., Squires, C. & Uhler, C. DCI: learning causal differences between gene regulatory networks. Bioinformatics 37, 3067–3069 (2021).
Tam, G. H. F., Chang, C. & Hung, Y. S. Gene regulatory network discovery using pairwise Granger causality. IET Syst. Biol. 7, 195–204 (2013).
Ke, N. R. et al. DiscoGen: learning to discover gene regulatory networks. Preprint at bioRxiv https://doi.org/10.1101/2023.04.11.536361 (2023).
Badia-I-Mompel, P. et al. Gene regulatory network inference in the era of single-cell multi-omics. Nat. Rev. Genet. 24, 739–754 (2023).
Fleck, J. S. et al. Inferring and perturbing cell fate regulomes in human brain organoids. Nature 621, 365–372 (2023).
Bravo González-Blas, C. et al. SCENIC+: single-cell multiomic inference of enhancers and gene regulatory networks. Nat. Methods 20, 1355–1367 (2023).
Santos-Zavaleta, A. et al. RegulonDB v 10.5: tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 47, D212–D220 (2019).
Peters, J., Janzing, D. & Scholkopf, B. Elements of Causal Inference: Foundations and Learning Algorithms (MIT Press, 2017).
Lopez, R., Hutter, J.-C., Pritchard, J. & Regev, A. Large-scale differentiable causal discovery of factor graphs. Neural Inf. Process. Syst. abs/2206.07824, 19290–19303 (2022).
Chevalley, M., Roohani, Y., Mehrjou, A., Leskovec, J. & Schwab, P. CausalBench: a large-scale benchmark for network inference from single-cell perturbation data. Preprint at https://arxiv.org/abs/2210.17283 (2022).
Wang, Y., Solus, L., Yang, K. D. & Uhler, C. Permutation-based causal inference algorithms with interventions. Neural Inf. Process. Syst. 30, 5822–5831 (2017).
Aliee, H., Kapl, F., Hediyeh-Zadeh, S. & Theis, F. J. Conditionally invariant representation learning for disentangling cellular heterogeneity. Preprint at https://arxiv.org/abs/2307.00558 (2023).
Levine, M. & Davidson, E. H. Gene regulatory networks for development. Proc. Natl Acad. Sci. USA 102, 4936–4942 (2005).
Lazar, N. H. et al. High-resolution genome-wide mapping of chromosome-arm-scale truncations induced by CRISPR–Cas9 editing. Nat. Genet. 56, 1482–1493 (2024).
Hsu, P. D. et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832 (2013).
Adikusuma, F. et al. Large deletions induced by Cas9 cleavage. Nature 560, E8–E9 (2018).
Tsuchida, C. A. et al. Mitigation of chromosome loss in clinical CRISPR–Cas9-engineered T cells. Cell 186, 4567–4582 (2023).
Papalexi, E. et al. Characterizing the molecular regulation of inhibitory immune checkpoints with multimodal single-cell screens. Nat. Genet. 53, 322–331 (2021).
Bunne, C. et al. Learning single-cell perturbation responses using neural optimal transport. Nat. Methods 20, 1759–1768 (2023).
Heumos, L. et al. Pertpy: an end-to-end framework for perturbation analysis. Preprint at bioRxiv https://doi.org/10.1101/2024.08.04.606516 (2024).
Dixit, A. et al. Perturb-seq: dissecting molecular circuits with scalable single-cell RNA profiling of pooled genetic screens. Cell 167, 1853–1866 (2016).
Norman, T. M. et al. Exploring genetic interaction manifolds constructed from rich single-cell phenotypes. Science 365, 786–793 (2019).
Replogle, J. M. et al. Mapping information-rich genotype–phenotype landscapes with genome-scale Perturb-seq. Cell 185, 2559–2575 (2022).
Lotfollahi, M., Wolf, F. A. & Theis, F. J. scGen predicts single-cell perturbation responses. Nat. Methods 16, 715–721 (2019).
Tran, H. T. N. et al. A benchmark of batch-effect correction methods for single-cell RNA sequencing data. Genome Biol. 21, 12 (2020).
Tung, P.-Y. et al. Batch effects and the effective design of single-cell gene expression studies. Sci. Rep. 7, 39921 (2017).
Rainforth, T., Foster, A., Ivanova, D. R. & Bickford Smith, F. Modern Bayesian experimental design. Stat. Sci. 39, 100–114 (2024).
Jain, M. et al. GFlowNets for AI-driven scientific discovery. Digit. Discov. 2, 557–577 (2023).
Williams, C. & Rasmussen, C. Gaussian processes for regression. In Advances in Neural Information Processing Systems (eds Touretzky, D. et al.) 514–520 (MIT Press, 1995).
Gal, Y. & Ghahramani, Z. Dropout as a Bayesian approximation: representing model uncertainty in deep learning. ICML 48, 1050–1059 (2015).
Lakshminarayanan, B., Pritzel, A. & Blundell, C. Simple and scalable predictive uncertainty estimation using deep ensembles. In Advances in Neural Information Processing Systems 405, 6402–6413 (2017).
Lahlou, S. et al. DEUP: direct epistemic uncertainty prediction. Trans. Mach. Learn. Res. (in the press).
Ke, N. R. et al. Learning neural causal models from unknown interventions. Preprint at https://arxiv.org/abs/1910.01075 (2019).
Deleu, T. et al. Bayesian structure learning with generative flow networks. In Proceedings of the Thirty-Eighth Conference on Uncertainty in Artificial Intelligence 518–528 (2022).
Močkus, J. On Bayesian methods for seeking the extremum. In Optimization Techniques IFIP Technical Conference Novosibirsk (ed. Marchuk, G. I.) 400–404 (Springer, 1975).
Toth, C. et al. Active Bayesian causal inference. Adv. Neural Inf. Proc. Syst. 35, 16261–16275 (2022).
Scherrer, N. et al. Learning neural causal models with active interventions. Preprint at https://arxiv.org/abs/2109.02429 (2021).
Smith, J. S., Nebgen, B., Lubbers, N., Isayev, O. & Roitberg, A. E. Less is more: sampling chemical space with active learning. J. Chem. Phys. 148, 241733 (2018).
Tran, K. et al. Computational catalyst discovery: active classification through myopic multiscale sampling. J. Chem. Phys. 154, 124118 (2021).
Kim, S. et al. Deep learning for Bayesian optimization of scientific problems with high-dimensional structure. Preprint at https://arxiv.org/abs/2104.11667 (2021).
Bertin, P. et al. RECOVER identifies synergistic drug combinations in vitro through sequential model optimization. Cell Rep. Methods 3, 100599 (2023).
Tosh, C. et al. A Bayesian active learning platform for scalable combination drug screens. Nat. Commun. 16, 156 (2025).
Szklarczyk, D. et al. The STRING database in 2023: protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2022).
Türei, D., Korcsmáros, T. & Saez-Rodriguez, J. OmniPath: guidelines and gateway for literature-curated signaling pathway resources. Nat. Methods 13, 966–967 (2016).
Lobentanzer, S. et al. Democratizing knowledge representation with BioCypher. Nat. Biotechnol. 41, 1056–1059 (2023).
Bertin, P. et al. Analysis of gene interaction graphs as prior knowledge for machine learning models. Preprint at https://arxiv.org/abs/1905.02295 (2019).
Lopez, R., Regier, J., Cole, M. B., Jordan, M. I. & Yosef, N. Deep generative modeling for single-cell transcriptomics. Nat. Methods 15, 1053–1058 (2018).
Stein-O’Brien, G. L. et al. Enter the matrix: factorization uncovers knowledge from omics. Trends Genet. 34, 790–805 (2018).
Piran, Z., Cohen, N., Hoshen, Y. & Nitzan, M. Disentanglement of single-cell data with biolord. Nat. Biotechnol. 42, 1678–1683 (2024).
Lotfollahi, M. et al. Predicting cellular responses to complex perturbations in high-throughput screens. Mol. Syst. Biol. 19, e11517 (2023).
Schölkopf, B. et al. Toward causal representation learning. Proc. IEEE 109, 612–634 (2021).
Ahuja, K., Mahajan, D., Wang, Y. & Bengio, Y. Interventional causal representation learning. Proc. 40th Intl Conf. Mach. Learn. 202, 372–407 (2023).
Varici, B., Acarturk, E., Shanmugam, K., Kumar, A. & Tajer, A. Score-based causal representation learning with interventions. Preprint at https://arxiv.org/abs/2301.08230 (2023).
Michael, B. & Karaletsos, T. Modelling cellular perturbations with the sparse additive mechanism shift variational autoencoder. Adv. Neural Inf. Proc. Syst. 36, 1–12 (2023).
Lotfollahi, M. et al. Biologically informed deep learning to query gene programs in single-cell atlases. Nat. Cell Biol. 25, 337–350 (2023).
Lopez, R. et al. Learning causal representations of single cells via sparse mechanism shift modeling. Proc. Mach. Learn. Res. 213, 1–30 (2023).
Kartik, A., Hartford, J. S. & Bengio, Y. Weakly supervised representation learning with sparse perturbations. Adv. Neural Inf. Process. Syst. 35, 15516–15528 (2022).
Peters, J., Bauer, S. & Pfister, N. in Causal Models for Dynamical Systems. Probabilistic and Causal Inference: The Works of Judea Pearl 1st edn. 671–690 (Association for Computing Machinery, 2022).
Haghverdi, L., Büttner, M., Wolf, F. A., Buettner, F. & Theis, F. J. Diffusion pseudotime robustly reconstructs lineage branching. Nat. Methods 13, 845–848 (2016).
Saelens, W., Cannoodt, R., Todorov, H. & Saeys, Y. A comparison of single-cell trajectory inference methods. Nat. Biotechnol. 37, 547–554 (2019).
Moon, K. R. et al. Manifold learning-based methods for analyzing single-cell RNA-sequencing data. Curr. Opin. Syst. Biol. 7, 36–46 (2018).
Aliee, H., Theis, F. J. & Kilbertus, N. Beyond predictions in neural ODEs: identification and interventions. Preprint at https://arxiv.org/abs/2106.12430 (2021).
Hananeh, A. et al. Sparsity in continuous-depth neural networks. Adv. Neural Inf. Process. Syst. 35, 901–914 (2022).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
Tong, A. et al. Trajectorynet: A dynamic optimal transport network for modeling cellular dynamics. Internatl Conf. Mach. Learn. http://proceedings.mlr.press/v119/tong20a/tong20a-supp.pdf (PMLR, 2020).
Schiebinger, G. et al. Optimal-transport analysis of single-cell gene expression identifies developmental trajectories in reprogramming. Cell 176, 928–943 (2019).
Eyring, L. V. et al. Modeling single-cell dynamics using unbalanced parameterized Monge maps. Preprint at bioRxiv https://doi.org/10.1101/2022.10.04.510766 (2022).
Wu, Y. et al. PerturBench: benchmarking machine learning models for cellular perturbation analysis. Preprint at https://arxiv.org/abs/2408.10609 (2024).
Csendes, G., Szalay, K. Z. & Szalai, B. Benchmarking a foundational cell model for post-perturbation RNAseq prediction. Preprint at bioRxiv https://doi.org/10.1101/2024.09.30.615843 (2024).
Mehrjou, A. et al. GeneDisco: a benchmark for experimental design in drug discovery. Preprint at https://arxiv.org/abs/2110.11875 (2021).
Metzner, E., Southard, K. M. & Norman, T. M. Multiome Perturb-seq unlocks scalable discovery of integrated perturbation effects on the transcriptome and epigenome. Cell Syst. 16, 101161 (2025).
Sethuraman, M. G. et al. NODAGS-Flow: nonlinear cyclic causal structure learning. In International Conference on Artificial Intelligence and Statistics (eds Ruiz, F. et al.) 6371–6387 (PMLR, 2023).
Nguyen, T., Tong, A., Madan, K., Bengio, Y. & Liu, D. Causal inference in gene regulatory networks with GFlowNet: towards scalability in large systems. Preprint at https://arxiv.org/abs/2310.03579 (2023).
Tung, K.-F., Pan, C.-Y., Chen, C.-H. & Lin, W.-C. Top-ranked expressed gene transcripts of human protein-coding genes investigated with GTEx dataset. Sci. Rep. 10, 16245 (2020).
Dhamija, S. & Menon, M. B. Non-coding transcript variants of protein-coding genes — what are they good for? RNA Biol. 15, 1025–1031 (2018).
Aebersold, R. et al. How many human proteoforms are there? Nat. Chem. Biol. 14, 206–214 (2018).
Dubey, A. et al. The Llama 3 herd of models. Preprint at https://arxiv.org/abs/2407.21783 (2024).
Gavrilov, A. A. et al. Studying RNA–DNA interactome by Red-C identifies noncoding RNAs associated with various chromatin types and reveals transcription dynamics. Nucleic Acids Res. 48, 6699–6714 (2020).
Noh, J. Y. et al. CCIDB: a manually curated cell–cell interaction database with cell context information. Database 2023, baad057 (2023).
Pearce, A. C. et al. Vav1 and Vav3 have critical but redundant roles in mediating platelet activation by collagen. J. Biol. Chem. 279, 53955–53962 (2004).
Author information
Authors and Affiliations
Contributions
A.T.-L. and P.B. conceptualized the work. P.B., A.T.-L. and H.A. wrote the original draft and created the figures. S.B. provided useful feedback. All authors critically reviewed and edited the manuscript. F.J.T., Y.B. and H.A. supervised the project.
Corresponding authors
Ethics declarations
Competing interests
Y.B. is an advisor to Recursion Pharmaceuticals. F.J.T. consults for Immunai, CytoReason, Cellarity, BioTuring and Genbio.AI and has an ownership interest in Dermagnostix and Cellarity. All other authors declare no competing interests.
Peer review
Peer review information
Nature Genetics thanks Patrick Schwab and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tejada-Lapuerta, A., Bertin, P., Bauer, S. et al. Causal machine learning for single-cell genomics. Nat Genet 57, 797–808 (2025). https://doi.org/10.1038/s41588-025-02124-2
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02124-2
This article is cited by
-
Interpretation, extrapolation and perturbation of single cells
Nature Reviews Genetics (2026)
-
Reference-guided computational framework identifies microenvironment metabolic subtypes and targets using pan-cancer single-cell datasets
Genome Medicine (2025)
-
In silico biological discovery with large perturbation models
Nature Computational Science (2025)