Fig. 1: scTF-seq design and the corresponding TF overexpression atlas.
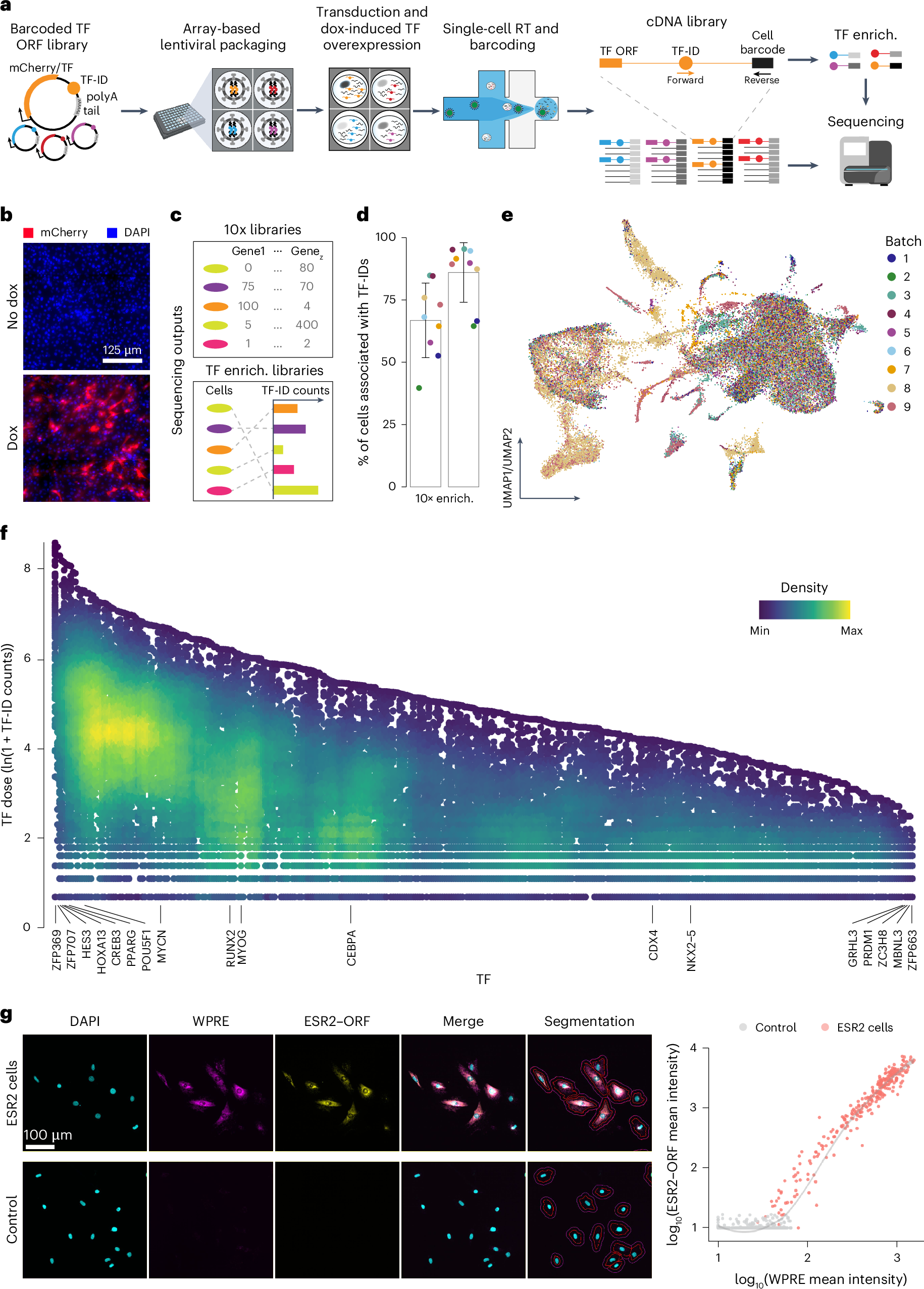
a, Schematic of the scTF-seq workflow. TF-ID, a unique barcode designed for mCherry (as control) or each individual TF; forward and reverse, primers to enrich TF-IDs. The arrayed screening schematic is created with BioRender.com. b, Fluorescence images of mCherry (red) and nuclei (DAPI, blue) in C3H10T1/2 cells treated without (no dox) or with doxycycline (dox). Representative images of more than three independent experiments. Scale bar = 125 μm. c, Schematic of the sequencing outputs of scTF-seq—count matrices of gene expression in 10x libraries (top) and ectopic TF-ID expression in TF-enrichment libraries (bottom) for each sequenced cell. d, Percentage of cell barcodes associated with TF-IDs in 10x or TF-enrichment libraries. Colors represent nine independent scTF-seq experiments (also referred to as ‘batches’, see color legend in e). Error bars represent the mean ± s.d. e, UMAP of scTF-seq data involving 45,987 cells and 384 TFs after quality control and preprocessing (referred to as ‘TF atlas’). Colors represent batches. f, Natural log-transformed TF expression levels (TF dose) in cells overexpressing individual TFs. Colors represent cell density (number of neighbors) after randomly sampling up to 500 cells for each TF. g, Left: RNAscope images for DAPI, WPRE (proxy for TF dose), ESR2–ORF in ESR2 (top) and control (bottom) cells. All fluorescence channels were merged for cell segmentation, indicated by the red (cell boundary) and purple (expanded cell boundary) outlines. Representative images of two independent experiments. Scale bar = 100 μm. Right: single-cell RNAscope quantification showing the log-normalized mean intensity of WPRE versus ESR2–ORF in control and ESR2 cells. Fitted model = LOESS (Extended Data Figs. 1 and 2). RT, reverse transcription; LOESS, locally estimated scatterplot smoothing; UMAP, uniform manifold approximation and projection; enrich., enrichment.
