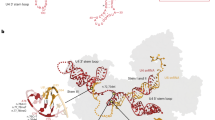
Abstract
Small nuclear RNA (snRNA) genes represent a class of non-protein-coding genes involved in the processing of pre-mRNAs of intron-containing genes. The human genome contains approximately 2,000 snRNA genes; the majority are pseudogenes, and only a small fraction are functional. These snRNAs undergo extensive post-transcriptional modifications, and, together with proteins and other snRNAs, form small nuclear ribonucleoproteins, which are components of the spliceosome. This Review discusses high-impact variants in 12 snRNA genes that cause Mendelian disorders with either autosomal dominant or recessive inheritance patterns. The associated phenotypes include mainly neurodevelopmental delay, developmental abnormalities and retinitis pigmentosa. The presumed consequences of these variants are presented on the basis of previous functional characterization of the corresponding snRNAs. It is anticipated that the understanding of both Mendelian and complex traits due to snRNAs will increase the diagnostic potential, partially explain penetrance and provide more therapeutic options.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Holley, R. W. et al. Structure of a ribonucleic acid. Science 147, 1462–1465 (1965).
Henderson, A. S., Warburton, D. & Atwood, K. C. Location of ribosomal DNA in the human chromosome complement. Proc. Natl Acad. Sci. USA 69, 3394–3398 (1972).
Brannan, C. I., Dees, E. C., Ingram, R. S. & Tilghman, S. M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 10, 28–36 (1990).
Brown, C. J. et al. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349, 38–44 (1991).
Lee, R. C., Feinbaum, R. L. & Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75, 843–854 (1993).
Wightman, B., Ha, I. & Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 75, 855–862 (1993).
Lander, E. S. et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001).
Venter, J. C. et al. The sequence of the human genome. Science 291, 1304–1351 (2001).
Nurk, S. et al. The complete sequence of a human genome. Science 376, 44–53 (2022).
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
Mudge, J. M. et al. GENCODE 2025: reference gene annotation for human and mouse. Nucleic Acids Res. 53, D966–D975 (2025).
Nakashima, T., et al. Diversity of U1 small nuclear RNAs and diagnostic methods for their mutations. Cancer Sci. 116, 2270–2280 (2025).
Nemeth, K., Bayraktar, R., Ferracin, M. & Calin, G. A. Non-coding RNAs in disease: from mechanisms to therapeutics. Nat. Rev. Genet. 25, 211–232 (2024).
Matera, A. G., Terns, R. M. & Terns, M. P. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8, 209–220 (2007).
Lerner, M. R. & Steitz, J. A. Antibodies to small nuclear RNAs complexed with proteins are produced by patients with systemic lupus erythematosus. Proc. Natl Acad. Sci. USA 76, 5495–5499 (1979).
Hodnett, J. L. & Busch, H. Isolation and characterization of uridylic acid-rich 7 S ribonucleic acid of rat liver nuclei. J. Biol. Chem. 243, 6334–6342 (1968).
Berget, S. M., Moore, C. & Sharp, P. A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl Acad. Sci. USA 74, 3171–3175 (1977).
Chow, L. T., Gelinas, R. E., Broker, T. R. & Roberts, R. J. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12, 1–8 (1977).
Shi, Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat. Rev. Mol. Cell Biol. 18, 655–670 (2017).
Will, C. L. & Luhrmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 3, a003707 (2011).
Verma, B., Akinyi, M. V., Norppa, A. J. & Frilander, M. J. Minor spliceosome and disease. Semin. Cell Dev. Biol. 79, 103–112 (2018).
Wilkinson, M. E., Charenton, C. & Nagai, K. RNA splicing by the spliceosome. Annu. Rev. Biochem. 89, 359–388 (2020).
Sharp, P. A. & Burge, C. B. Classification of introns: U2-type or U12-type. Cell 91, 875–879 (1997).
Jouravleva, K. & Zamore, P. D. A guide to the biogenesis and functions of endogenous small non-coding RNAs in animals. Nat. Rev. Mol. Cell Biol. 26, 347–370 (2025).
Matera, A. G. & Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 15, 108–121 (2014).
Wan, R., Bai, R., Zhan, X. & Shi, Y. How is precursor messenger RNA spliced by the spliceosome? Annu. Rev. Biochem. 89, 333–358 (2020).
Mabin, J. W., Lewis, P. W., Brow, D. A. & Dvinge, H. Human spliceosomal snRNA sequence variants generate variant spliceosomes. RNA 27, 1186–1203 (2021).
Sontheimer, E. J. & Steitz, J. A. Three novel functional variants of human U5 small nuclear RNA. Mol. Cell. Biol. 12, 734–746 (1992).
Prasetyo, N. K. & Gardner, P. P. Assessing the robustness of human ncRNA notation at HGNC. Preprint at bioRxiv https://doi.org/10.1101/2024.12.08.627405 (2024).
Chen, Y. et al. De novo variants in the RNU4-2 snRNA cause a frequent neurodevelopmental syndrome. Nature 632, 832–840 (2024).
Greene, D. et al. Mutations in the U4 snRNA gene RNU4-2 cause one of the most prevalent monogenic neurodevelopmental disorders. Nat. Med. 30, 2165–2169 (2024).
Edery, P. et al. Association of TALS developmental disorder with defect in minor splicing component U4atac snRNA. Science 332, 240–243 (2011).
He, H. et al. Mutations in U4atac snRNA, a component of the minor spliceosome, in the developmental disorder MOPD I. Science 332, 238–240 (2011).
Barbour, K. et al. The face and features of RNU4-2: a new, common, recognizable, yet hidden neurodevelopmental disorder. Pediatr. Neurol. 161, 188–193 (2024).
Valenzuela, I. et al. Deep phenotyping of 11 individuals with pathogenic variants in RNU4-2 reveals a clinically recognizable syndrome. Genet. Med. 26, 101288 (2024).
Charenton, C., Wilkinson, M. E. & Nagai, K. Mechanism of 5′ splice site transfer for human spliceosome activation. Science 364, 362–367 (2019).
Nava, C. et al. Dominant variants in major spliceosome U4 and U5 small nuclear RNA genes cause neurodevelopmental disorders through splicing disruption. Nat. Genet. 57, 1374–1388 (2025).
Schot, R., Ferraro, F., Geeven, G., Diderich, K. E. M. & Barakat, T. S. Re-analysis of whole genome sequencing ends a diagnostic odyssey: case report of an RNU4-2 related neurodevelopmental disorder. Clin. Genet. 106, 512–517 (2024).
De Jonghe, J. et al. Saturation genome editing of RNU4-2 reveals distinct dominant and recessive neurodevelopmental disorders. Preprint at medRxiv https://doi.org/10.1101/2025.04.08.25325442 (2025).
Bertoli-Avella, A. M. RNU4-2 monoallelic variants as a leading cause of syndromic neurodevelopmental disorder, including in patients with parental consanguinity. J. Med. Genet. 62, 536–539 (2025).
Bruselles, A. et al. Expanding the mutational spectrum of ReNU syndrome: insights into 5′ stem-loop variants. Eur. J. Hum. Genet. 33, 432–440 (2025).
Okamoto, N. A clinical study of nine patients with ReNU syndrome. Am. J. Med. Genet. A 197, e64151 (2025).
Quinodoz, M. et al. De novo and inherited dominant variants in U4 and U6 snRNAs cause retinitis pigmentosa. Preprint at medRxiv https://doi.org/10.1101/2025.01.06.24317169 (2025).
Perea-Romero, I. et al. Genetic landscape of 6089 inherited retinal dystrophies affected cases in Spain and their therapeutic and extended epidemiological implications. Sci. Rep. 11, 1526 (2021).
Hardin, J. W., Warnasooriya, C., Kondo, Y., Nagai, K. & Rueda, D. Assembly and dynamics of the U4/U6 di-snRNP by single-molecule FRET. Nucleic Acids Res. 43, 10963–10974 (2015).
Liu, S. et al. Binding of the human Prp31 Nop domain to a composite RNA–protein platform in U4 snRNP. Science 316, 115–120 (2007).
Rius, R. et al. Biallelic variants in the non-coding RNA gene RNU4-2 cause a recessive neurodevelopmental syndrome with distinct white matter changes. Preprint at medRxiv https://doi.org/10.1101/2025.08.13.25333306 (2025).
Greene, D. et al. Mutations in the small nuclear RNA gene RNU2-2 cause a severe neurodevelopmental disorder with prominent epilepsy. Nat. Genet. 57, 1367–1373 (2025).
Bousquets-Munoz, P., et al. PanCancer analysis of somatic mutations in repetitive regions reveals recurrent mutations in snRNA U2. NPJ Genom. Med. 7, 19 (2022).
Xie, J., Wang, L. & Lin, R. J. Variations of intronic branchpoint motif: identification and functional implications in splicing and disease. Commun. Biol. 6, 1142 (2023).
100,000 Genomes Project Pilot Investigators et al. 100,000 Genomes Pilot on rare-disease diagnosis in health care — preliminary report. N. Engl. J. Med. 385, 1868–1880 (2021).
Jonsson, H. et al. Parental influence on human germline de novo mutations in 1,548 trios from Iceland. Nature 549, 519–522 (2017).
Jackson, A. et al. Analysis of R-loop forming regions identifies RNU2-2 and RNU5B-1 as neurodevelopmental disorder genes. Nat. Genet. 57, 1362–1366 (2025).
Jia, Y., Mu, J. C. & Ackerman, S. L. Mutation of a U2 snRNA gene causes global disruption of alternative splicing and neurodegeneration. Cell 148, 296–308 (2012).
Greene, D. et al. Biallelic variants in RNU2-2 cause the most prevalent known recessive neurodevelopmental disorder. Preprint at medRxiv https://doi.org/10.1101/2025.08.26.25334179 (2025).
Jackson, A. et al. Biallelic variants in RNU2-2 cause a remarkably frequent developmental epileptic encephalopathy. Preprint at medRxiv https://doi.org/10.1101/2025.09.02.25334957 (2025).
Leitao, E. et al. Systematic analysis of snRNA genes reveals frequent RNU2-2 variants in dominant and recessive developmental and epileptic encephalopathies. Preprint at medRxiv https://doi.org/10.1101/2025.09.02.25334923 (2025).
Benoit-Pilven, C., et al. Clinical interpretation of variants identified in RNU4ATAC, a non-coding spliceosomal gene. PLoS ONE 15, e0235655 (2020).
Norppa, A. J., Shcherbii, M. V. & Frilander, M. J. Connecting genotype and phenotype in minor spliceosome diseases. RNA 31, 284–299 (2025).
Duker, A. et al. RNU4atac-opathy. In GeneReviews (eds Adam, M. P. et al.) (Univ. Washington, 1993).
Merico, D. et al. Compound heterozygous mutations in the noncoding RNU4ATAC cause Roifman syndrome by disrupting minor intron splicing. Nat. Commun. 6, 8718 (2015).
Heremans, J. et al. Abnormal differentiation of B cells and megakaryocytes in patients with Roifman syndrome. J. Allergy Clin. Immunol. 142, 630–646 (2018).
Khatri, D. et al. Deficiency of the minor spliceosome component U4atac snRNA secondarily results in ciliary defects in human and zebrafish. Proc. Natl Acad. Sci. USA 120, e2102569120 (2023).
Hagiwara, H. et al. Immunodeficiency in a patient with microcephalic osteodysplastic primordial dwarfism type I as compared to Roifman syndrome. Brain Dev. 43, 337–342 (2021).
Arriaga, M. T. et al. Transcriptome-wide outlier approach identifies individuals with minor spliceopathies. Am. J. Hum. Genet. 112, 2458–2475 (2025).
Elsaid, M. F. et al. Mutation in noncoding RNA RNU12 causes early onset cerebellar ataxia. Ann. Neurol. 81, 68–78 (2017).
Norppa, A. J. & Frilander, M. J. The integrity of the U12 snRNA 3′ stem-loop is necessary for its overall stability. Nucleic Acids Res. 49, 2835–2847 (2021).
Konig, H., Matter, N., Bader, R., Thiele, W. & Muller, F. Splicing segregation: the minor spliceosome acts outside the nucleus and controls cell proliferation. Cell 131, 718–729 (2007).
Xing, C. et al. Biallelic variants in RNU12 cause CDAGS syndrome. Hum. Mutat. 42, 1042–1052 (2021).
Huang, Q., Jacobson, M. R. & Pederson, T. 3′ processing of human pre-U2 small nuclear RNA: a base-pairing interaction between the 3′ extension of the precursor and an internal region. Mol. Cell. Biol. 17, 7178–7185 (1997).
Gadgil, A. & Raczynska, K. D. U7 snRNA: a tool for gene therapy. J. Gene Med. 23, e3321 (2021).
Grimm, C., Stefanovic, B. & Schumperli, D. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 12, 1229–1238 (1993).
Schaufele, F., Gilmartin, G. M., Bannwarth, W. & Birnstiel, M. L. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3′ end of H3 messenger RNA. Nature 323, 777–781 (1986).
Kolev, N. G. & Steitz, J. A. In vivo assembly of functional U7 snRNP requires RNA backbone flexibility within the Sm-binding site. Nat. Struct. Mol. Biol. 13, 347–353 (2006).
Marquis, J. et al. Spinal muscular atrophy: SMN2 pre-mRNA splicing corrected by a U7 snRNA derivative carrying a splicing enhancer sequence. Mol. Ther. 15, 1479–1486 (2007).
Meyer, K. et al. Rescue of a severe mouse model for spinal muscular atrophy by U7 snRNA-mediated splicing modulation. Hum. Mol. Genet. 18, 546–555 (2009).
Uggenti, C. et al. cGAS-mediated induction of type I interferon due to inborn errors of histone pre-mRNA processing. Nat. Genet. 52, 1364–1372 (2020).
Naesens, L. et al. Mutations in RNU7-1 weaken secondary RNA structure, induce MCP-1 and CXCL10 in CSF, and result in Aicardi–Goutieres syndrome with severe end-organ involvement. J. Clin. Immunol. 42, 962–974 (2022).
MacArthur, D. G. et al. Guidelines for investigating causality of sequence variants in human disease. Nature 508, 469–476 (2014).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 17, 405–424 (2015).
Kong, A. et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 488, 471–475 (2012).
Porubsky, D., et al. Human de novo mutation rates from a four-generation pedigree reference. Nature 643, 427–436 (2025).
Didychuk, A. L., Butcher, S. E. & Brow, D. A. The life of U6 small nuclear RNA, from cradle to grave. RNA 24, 437–460 (2018).
Becker, D. et al. Nuclear pre-snRNA export is an essential quality assurance mechanism for functional spliceosomes. Cell Rep. 27, 3199–3214 (2019).
Schumperli, D. & Pillai, R. S. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. 61, 2560–2570 (2004).
Pillai, R. S. et al. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 17, 2321–2333 (2003).
Acknowledgements
I thank the ChildCare Foundation for support and colleagues at the Department of Genetic Medicine and Development of the Medical Faculty of the University of Geneva and Medigenome, the Swiss Institute of Genomic Medicine, for discussions. I thank C. Rivolta, D.A. Brow and R.S. Pillai for constructive comments on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
S.E.A. is a cofounder and the CEO of Medigenome, Swiss Institute of Genomic Medicine, and is also a member of the scientific advisory board of the Imagine Institute of the Necker Hospital in Paris, France.
Peer review
Peer review information
Nature Genetics thanks the anonymous reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Tables 1 and 2 and Figs. 1–3
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Antonarakis, S.E. Small nuclear RNA genes in Mendelian disorders. Nat Genet 58, 28–38 (2026). https://doi.org/10.1038/s41588-025-02440-7
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41588-025-02440-7