Abstract
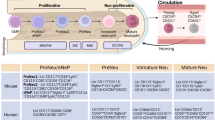
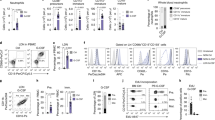
Neutrophils are formidable defenders. Their vast numbers, constant production, high cytotoxicity and capacity to produce extracellular traps, underlie their ability to efficiently protect in a microorganism-rich world. However, neutrophils are much more than immune sentinels, as evidenced by the expanding repertoire of functions discovered in the context of tissue homeostasis, regeneration or chronic pathologies. In this Perspective, we discuss general functional features of the neutrophil compartment that may be relevant in most, if not all, physiological scenarios in which they participate, including specialization in naïve tissues, transcriptional noise in the bloodstream as a potential strategy for diversification and functional bias in inflammatory sites. We intentionally present the reader with more questions than answers and propose models and approaches that we hope will shed new light onto the biology of these fascinating cells and spark new directions of research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Burn, G. L., Foti, A., Marsman, G., Patel, D. F. & Zychlinsky, A. The neutrophil. Immunity 54, 1377–1391 (2021).
Ley, K. et al. Neutrophils: new insights and open questions. Sci. Immunol. 3, eaat4579 (2018).
Aroca-Crevillén, A., Adrover, J. M. & Hidalgo, A. Circadian features of neutrophil biology. Front. Immunol. 11, 576 (2020).
Dancey, J. T., Deubelbeiss, K. A., Harker, L. A. & Finch, C. A. Neutrophil kinetics in man. J. Clin. Invest. 58, 705–715 (1976).
Lahoz-Beneytez, J. et al. Human neutrophil kinetics: modeling of stable isotope labeling data supports short blood neutrophil half-lives. Blood 127, 3431–3438 (2016).
Pillay, J. et al. In vivo labeling with 2H2O reveals a human neutrophil lifespan of 5.4 days. Blood 116, 625–627 (2010).
Ai, Z. & Udalova, I. A. Transcriptional regulation of neutrophil differentiation and function during inflammation. J. Leukoc. Biol. 107, 419–430 (2020).
Grassi, L. et al. Dynamics of transcription regulation in human bone marrow myeloid differentiation to mature blood neutrophils. Cell Rep. 24, 2784–2794 (2018).
Theilgaard-Mönch, K. et al. The transcriptional program of terminal granulocytic differentiation. Blood 105, 1785–1796 (2005).
Ballesteros, I. et al. Co-option of neutrophil fates by tissue environments. Cell 183, 1282–1297 (2020).
Casanova‑Acebes, M. et al. Neutrophils instruct homeostatic and pathological states in naive tissues. J. Exp. Med. 215, 2778–2795 (2018).
Khoyratty, T. E. et al. Distinct transcription factor networks control neutrophil-driven inflammation. Nat. Immunol. 22, 1093–1106 (2021).
Grieshaber-Bouyer, R. et al. The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat. Commun. 12, 2856 (2021).
Fine, N. et al. Distinct oral neutrophil subsets define health and periodontal disease states. J. Dent. Res. 95, 931–938 (2016).
Puga, I. et al. B cell-helper neutrophils stimulate the diversification and production of immunoglobulin in the marginal zone of the spleen. Nat. Immunol. 13, 170–180 (2012).
Xie, X. et al. Single-cell transcriptome profiling reveals neutrophil heterogeneity in homeostasis and infection. Nat. Immunol. 21, 1119–1133 (2020).
Hu, N. et al. Coexpression of CD177 and membrane proteinase 3 on neutrophils in antineutrophil cytoplasmic autoantibody-associated systemic vasculitis: anti-proteinase-3-mediated neutrophil activation is independent of the role of CD177-expressing neutrophils. Arthritis Rheum. 60, 1548–1557 (2009).
Silvestre-Roig, C., Hidalgo, A. & Soehnlein, O. Neutrophil heterogeneity: implications for homeostasis and pathogenesis. Blood 127, 2173–2181 (2016).
Verheugt, F. W. A., Borne, A. E. G. K., Noord-Bokhorsl, J. C., Elven, E. H. & Engelfriet, C. P. Serological, immunochemical and immuoncytological properties of granulocyte antibodies. Vox Sang. 35, 294–303 (1978).
Clemmensen, S. N. et al. Olfactomedin 4 defines a subset of human neutrophils. J. Leukoc. Biol. 91, 495–500 (2012).
Adrover, J. M. et al. Programmed ‘disarming’ of the neutrophil proteome reduces the magnitude of inflammation. Nat. Immunol. 21, 135–144 (2020).
Bowling, S. et al. An engineered CRISPR–Cas9 mouse line for simultaneous readout of lineage histories and gene expression profiles in single cells. Cells 181, 1410–1422 (2020).
Weinreb, C., Rodriguez-Fraticelli, A., Camargo, F. D. & Klein, A. M. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science 367, eaaw3381 (2020).
Scherer, A. K. et al. Predestined neutrophil heterogeneity in homeostasis varies in transcriptional and phenotypic response to Candida. Preprint at bioRxiv https://doi.org/10.1101/2022.11.01.514676 (2022).
Evrard, M. et al. Developmental analysis of bone marrow neutrophils reveals populations specialized in expansion, trafficking and effector functions. Immunity 48, 364–379 (2018).
Hedrick, C. C. & Malanchi, I. Neutrophils in cancer: heterogeneous and multifaceted.Nat. Rev. Immunol. 22, 173–187 (2022).
Manz, M. G. & Boettcher, S. Emergency granulopoiesis. Nat. Rev. Immunol. 14, 302–314 (2014).
van Grinsven, E. et al. Immature neutrophils released in acute inflammation exhibit efficient migration despite incomplete segmentation of the nucleus. J. Immunol. 202, 207–217 (2019).
Schulte-Schrepping, J. et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell 182, 1419–1440 (2020).
Combes, A. J. et al. Global absence and targeting of protective immune states in severe COVID-19. Nature 591, 124–130 (2021).
Wilk, A. J. et al. A single-cell atlas of the peripheral immune response in patients with severe COVID-19. Nat. Med. 26, 1070–1076 (2020).
Montaldo, E. et al. Cellular and transcriptional dynamics of human neutrophils at steady state and upon stress. Nat. Immunol. https://doi.org/10.1038/s41590-022-01311-1 (2022).
Engblom, C. et al. Osteoblasts remotely supply lung tumors with cancer-promoting SiglecFhigh neutrophils. Science 358, eaal5081 (2017).
Ryu, S. et al. Siglec-F–expressing neutrophils are essential for creating a profibrotic microenvironment in renal fibrosis. J. Clin. Invest. 132, e156876 (2022).
Christoffersson, G. & Phillipson, M. The neutrophil: one cell on many missions or many cells with different agendas? Cell Tissue Res. 371, 415–423 (2018).
Silvestre-Roig, C., Fridlender, Z. G., Glogauer, M. & Scapini, P. Neutrophil diversity in health and disease. Trends Immunol. 40, 565–583 (2019).
Christoffersson, G. et al. VEGF-A recruits a proangiogenic MMP-9-delivering neutrophil subset that induces angiogenesis in transplanted hypoxic tissue. Blood 120, 4653–4662 (2012).
Zhu, X. & Zhu, J. CD4 T helper cell subsets and related human immunological disorders. Int. J. Mol. Sci. 21, 8011 (2020).
Deniset, J. F. & Kubes, P. Neutrophil heterogeneity: bona fide subsets or polarization states? J. Leukoc. Biol. https://doi.org/10.1002/JLB.3RI0917-361R (2018).
Wigerblad, G. et al. Single-cell analysis reveals the range of transcriptional states of circulating human neutrophils. J. Immunol. 209, 772–782 (2022).
Ballesteros, I., Cerezo‐Wallis, D. & Hidalgo, A. Understanding NSCLC, one cell at a time. Cancer Cell https://doi.org/10.1038/nrc3060 (2022).
Adrover, J. M. et al. A neutrophil timer coordinates immune defense and vascular protection. Immunity 50, 390–402 (2019).
Zhang, D. et al. Neutrophil ageing is regulated by the microbiome. Nature 525, 528–532 (2015).
Iwafuchi-Doi, M. & Zaret, K. S. Pioneer transcription factors in cell reprogramming. Genes Dev. 28, 2679–2692 (2014).
Lavin, Y. et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 159, 1312–1326 (2014).
Ostuni, R. et al. Latent enhancers activated by stimulation in differentiated cells. Cell 152, 157–171 (2013).
Kalafati, L. et al. Innate immune training of granulopoiesis promotes anti-tumor activity. Cell 183, 771–785 (2020).
Moorlag, S. J. C. F. M. et al. BCG vaccination induces long-term functional reprogramming of human neutrophils. Cell Rep. 33, 108387 (2020).
Wu, Q. & Lucas, D. Resilient anatomy and local microplasticity of naïve and stress hematopoiesis. Preprint at bioRxiv https://doi.org/10.1101/2022.05.23.492315 (2020).
Zhang, J. et al. In situ mapping identifies distinct vascular niches for myelopoiesis. Nature 590, 457–462 (2020).
Osman, A. et al. Paired bone marrow and peripheral blood samples demonstrate lack of widespread dissemination of some CH clones. Blood Adv. 13, 269–274 (2022).
Nourshargh, S., Renshaw, S. A. & Imhof, B. A. Reverse migration of neutrophils: where, when, how and why. Trends Immunol. 37, 273–286 (2016).
Buckley, C. D. et al. Identification of a phenotypically and functionally distinct population of long-lived neutrophils in a model of reverse endothelial migration. J. Leukoc. Biol. 79, 303–311 (2006).
Kwok, I. et al. Combinatorial single-cell analyses of granulocyte–monocyte progenitor heterogeneity reveals an early uni-potent neutrophil progenitor. Immunity 53, 303–318 (2020).
Zhu, Y. P. et al. CyTOF mass cytometry reveals phenotypically distinct human blood neutrophil populations differentially correlated with melanoma stage. J. Immunother. Cancer 8, e000473 (2020).
Zhu, Y. P. et al. Identification of an early unipotent neutrophil progenitor with pro-tumoral activity in mouse and human bone marrow. Cell Rep. 24, 2329–2341 (2018).
Ecker, S. et al. Genome-wide analysis of differential transcriptional and epigenetic variability across human immune cell types. Genome Biol. 18, 18 (2017).
Naranbhai, V. et al. Genomic modulators of gene expression in human neutrophils. Nat. Commun. 6, 7545 (2015).
Rosales-Alvarez, R. E., Rettkowski, J., Herman, J. S., Cabezas-Wallscheid, N. & Grün, D. Gene expression noise dynamics unveil functional heterogeneity of ageing hematopoietic stem cells. Preprint at bioRxiv https://doi.org/10.1101/2022.08.04.502776 (2022).
Enge, M. et al. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell 171, 321–330 (2017).
Fraser, L. T. C. R., Dikdan, R. J., Dey, S., Singh, A. & Tyagi, S. Reduction in gene expression noise by targeted increase in accessibility at gene loci. Proc. Natl Acad. Sci. USA 118, e2018640118 (2021).
Xue, R. et al. Liver tumour immune microenvironment subtypes and neutrophil heterogeneity. Nature https://doi.org/10.1038/s41586-022-05400-x (2022).
Hackert, N. S., Radtke, F. A., Exner, T., Lorenz, H. & Grieshaber-Bouyer, R. Human and murine neutrophils share core transcriptional programs in both homeostatic and inflamed contexts. Preprint at bioRxiv https://doi.org/10.1101/2022.11.13.516246 (2022).
Simoni, Y., Chng, M. H. Y., Li, S., Fehlings, M. & Newell, E. W. Mass cytometry: a powerful tool for dissecting the immune landscape. Curr. Opin. Immunol. 51, 187–196 (2018).
Mihlan, M., Safaiyan, S., Stecher, M., Paterson, N. & Lämmermann, T. Surprises from intravital imaging of the innate immune response. Annu. Rev. Cell Dev. Biol. 38, 467–489 (2022).
Crainiciuc, G. et al. Behavioural immune landscapes of inflammation. Nature 601, 415–421 (2022).
Woodfin, A. et al. The junctional adhesion molecule JAM-C regulates polarized transendothelial migration of neutrophils in vivo. Nat. Immunol. 12, 761–769 (2011).
Pizzagalli, D. U. et al. Characterization of the dynamic behavior of neutrophils following influenza vaccination. Front. Immunol. 10, 2621 (2019).
Liew, P. X. & Kubes, P. The Neutrophil’s role during health and disease. Physiol. Rev. 99, 1223–1248 (2019).
Yipp, B. G. et al. The lung is a host defense niche for immediate neutrophil-mediated vascular protection. Sci. Immunol. 2, eaam8929 (2017).
Deniset, J. F., Surewaard, B. G., Lee, W. Y. & Kubes, P. Splenic Ly6Ghigh mature and Ly6Gint immature neutrophils contribute to eradication of S. pneumoniae. J. Exp. Med. 214, 1333–1350 (2017).
Ng, L. G. et al. Visualizing the neutrophil response to sterile tissue injury in mouse dermis reveals a three-phase cascade of events. J. Invest. Dermatol. https://doi.org/10.1038/jid.2011.179 (2011).
Beauvillain, C. et al. CCR7 is involved in the migration of neutrophils to lymph nodes. Blood 117, 1196–1204 (2011).
Devi, S. et al. Neutrophil mobilization via plerixaformediated CXCR4 inhibition arises from lung demargination and blockade of neutrophil homing to the bone marrow. J. Exp. Med. 210, 2321–2336 (2013).
He, W. et al. Circadian expression of migratory factors establishes lineage-specific signatures that guide the homing of leukocyte subsets to tissues. Immunity 49, 1175–1190 (2018).
Bratton, D. L. & Henson, P. M. Neutrophil clearance: when the party is over, clean-up begins. Trends Immunol. 32, 350–357 (2011).
A-Gonzalez, N. et al. Phagocytosis imprints heterogeneity in tissue-resident macrophages. J. Exp. Med. 214, 1281–1296 (2017).
Suratt, B. T. et al. Neutrophil maturation and activation determine anatomic site of clearance from circulation. Am. J. Physiol. Lung Cell. Mol. Physiol. 281, L913–L921 (2001).
Saverymuttu, S. H., Peters, A. M., Keshavarzian, A., Reavy, H. J. & Lavender, J. P. The kinetics of 111Indium distribution following injection of 111Indium labelled autologous granulocytes in man. Br. J. Haematol. 61, 675–685 (1985).
Furze, R. C. & Rankin, S. M. The role of the bone marrow in neutrophil clearance under homeostatic conditions in the mouse. FASEB J. 22, 3111–3119 (2008).
Guilliams, M., Thierry, G. R., Bonnardel, J. & Bajenoff, M. Establishment and maintenance of the macrophage niche. Immunity 52, 434–451 (2020).
Ginhoux, F. & Guilliams, M. Tissue-resident macrophage ontogeny and homeostasis. Immunity 44, 439–449 (2016).
Bowers, E. et al. Granulocyte-derived TNFα promotes vascular and hematopoietic regeneration in the bone marrow. Nat. Med. 24, 95–102 (2018).
Wang, J. et al. Visualizing the function and fate of neutrophils in sterile injury and repair. Science 358, 111–116 (2017).
Özcan, A. et al. CCR7-guided neutrophil redirection to skin-draining lymph nodes regulates cutaneous inflammation and infection. Sci. Immunol. 7, eabi9126 (2022).
Sreejit, G. et al. Retention of the NLRP3 inflammasome-primed neutrophils in the bone marrow is essential for myocardial infarction-induced granulopoiesis. Circulation 145, 31–44 (2022).
Casanova-Acebes, M. et al. Rhythmic modulation of the hematopoietic niche through neutrophil clearance. Cell 153, 1025–1035 (2013).
Calì, B. et al. Atypical CXCL12 signaling enhances neutrophil migration by modulating nuclear deformability. Sci. Signal. 15, eabk2552 (2022).
Cerezo‐Wallis, D. & Ballesteros, I. Neutrophils in cancer, a love–hate affair. FEBS J. 289, 3692–3703 (2022).
Coffelt, S. B., Wellenstein, M. D. & de Visser, K. E. Neutrophils in cancer: neutral no more. Nat. Rev. Cancer 16, 431–446 (2016).
Cuartero, M. I. et al. N2 neutrophils, novel players in brain inflammation after stroke: modulation by the PPARγ agonist rosiglitazone. Stroke 44, 3498–3508 (2013).
Ng, L. G., Ostuni, R. & Hidalgo, A. Heterogeneity of neutrophils. Nat. Rev. Immunol. 19, 255–265 (2019).
Vafadarnejad, E. et al. Dynamics of cardiac neutrophil diversity in murine myocardial infarction. Circ. Res. 127, e232–e249 (2020).
Sharma, A., Blériot, C., Currenti, J. & Ginhoux, F. Oncofetal reprogramming in tumour development and progression. Nat. Rev. Cancer 22, 593–602 (2022).
Bronte, V. et al. Recommendations for myeloid-derived suppressor cell nomenclature and characterization standards. Nat. Commun. 7, 12150 (2016).
Bachmaier, K. et al. Albumin nanoparticle endocytosing subset of neutrophils for precision therapeutic targeting of inflammatory tissue injury. ACS Nano 16, 4084–4101 (2022).
Raj, A., Peskin, C. S., Tranchina, D., Vargas, D. Y. & Tyagi, S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 4, 1707–1719 (2006).
Ibáñez-Solé, O., Ascensión, A. M., Araúzo-Bravo, M. J. & Izeta, A. Lack of evidence for increased transcriptional noise in aged tissues. eLife 11, e80380 (2022).
Acknowledgements
We thank members of the laboratory of A.H., P. Frenette, D. Lucas, L. Ng, G. Fernandez-Calvo, F. Sanchez-Cabo, M. Casanova-Acebes and J. M. Adrover for past and present discussion and inspiration on questions presented here. This manuscript has been possible through grants R01AI165661 from NIH/NIAD, RTI2018-095497-B-I00 from MCIN, HR17_00527 from Fundación La Caixa, the Transatlantic Network of Excellence (TNE-18CVD04) from the Leducq Foundation, and FET-OPEN (no. 861878) from the European Commission. M.P.-S. is supported by the EMBO ALTF (no. 1142-2020) long-term fellowship and from MICINN (RYC2021-033511-I). J.S. and I.B. are supported by fellowships from MICINN (PRE2019-089130 and RYC2020-029563-I, respectively). The CNIC is supported by the MCIN and the Pro CNIC Foundation and is a Severo Ochoa Center of Excellence (CEX2020-001041-S).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Immunology thanks Hongbo Luo and Renato Ostuni for their contribution to the peer review of this work. Primary Handling Editor: Ioana Visan, in collaboration with the Nature Immunology team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Palomino-Segura, M., Sicilia, J., Ballesteros, I. et al. Strategies of neutrophil diversification. Nat Immunol 24, 575–584 (2023). https://doi.org/10.1038/s41590-023-01452-x
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41590-023-01452-x
This article is cited by
-
Multimodal single-cell protein and RNA profiling unveils dysregulated immature neutrophil dynamics in gestational diabetes mellitus
Communications Biology (2026)
-
Immune dysfunction during S. aureus biofilm-associated implant infections: opportunities for novel therapeutic strategies
npj Biofilms and Microbiomes (2025)
-
Tumor cells that resist neutrophil anticancer cytotoxicity acquire a prometastatic and innate immune escape phenotype
Cellular & Molecular Immunology (2025)
-
The heterogeneity of neutrophils in cancer and its implication for therapeutic targeting
Nature Immunology (2025)
-
Targeting neutrophils for cancer therapy
Nature Reviews Drug Discovery (2025)