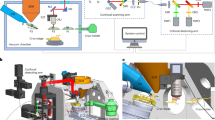
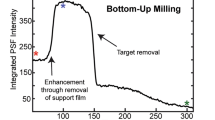
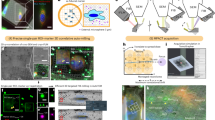
Abstract
Cryogenic electron microscopy and data processing enable the determination of structures of isolated macromolecules to near-atomic resolution. However, these data do not provide structural information in the cellular environment where macromolecules perform their native functions, and vital molecular interactions can be lost during the isolation process. Cryogenic focused ion beam (FIB) fabrication generates thin lamellae of cellular samples and tissues, enabling structural studies on the near-native cellular interior and its surroundings by cryogenic electron tomography (cryo-ET). Cellular cryo-ET benefits from the technological developments in electron microscopes, detectors and data processing, and more in situ structures are being obtained and at increasingly higher resolution. In this Review, we discuss recent studies employing cryo-ET on FIB-generated lamellae and the technological developments in ultrarapid sample freezing, FIB fabrication of lamellae, tomography, data processing and correlative light and electron microscopy that have enabled these studies. Finally, we explore the future of cryo-ET in terms of both methods development and biological application.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kuhlbrandt, W. The resolution revolution. Science 343, 1443–1444 (2014).
Nakane, T. et al. Single-particle cryo-EM at atomic resolution. Nature 587, 152–156 (2020).
Yip, K. M., Fischer, N., Paknia, E., Chari, A. & Stark, H. Atomic-resolution protein structure determination by cryo-EM. Nature 587, 157–161 (2020).
Dubochet, J. et al. Cryo-electron microscopy of vitrified specimens. Q. Rev. Biophys. 21, 129–228 (1988).
Medalia, O. Macromolecular architecture in eukaryotic cells visualized by cryoelectron tomography. Science 298, 1209–1213 (2002).
Dobro, M. J. et al. Uncharacterized bacterial structures revealed by electron cryotomography. J. Bacteriol. 199, 1–14 (2017).
Walz, J. et al. Tricorn protease exists as an icosahedral supermolecule in vivo. Mol. Cell 1, 59–65 (1997).
Wan, W. & Briggs, J. A. G. Cryo-electron tomography and subtomogram averaging. Methods Enzymol. 579, 329–367 (2016).
McDowall, A. W. et al. Electron microscopy of frozen hydrated sections of vitreous ice and vitrified biological samples. J. Microsc. 131, 1–9 (1983).
Al-Amoudi, A., Norlen, L. P. O. & Dubochet, J. Cryo-electron microscopy of vitreous sections of native biological cells and tissues. J. Struct. Biol. 148, 131–135 (2004).
Pierson, J., Ziese, U., Sani, M. & Peters, P. J. Exploring vitreous cryo-section-induced compression at the macromolecular level using electron cryo-tomography; 80S yeast ribosomes appear unaffected. J. Struct. Biol. 173, 345–349 (2011).
Marko, M., Hsieh, C., Schalek, R., Frank, J. & Mannella, C. Focused-ion-beam thinning of frozen-hydrated biological specimens for cryo-electron microscopy. Nat. Methods 4, 215–217 (2007). Key study in the development of cryo-FIB lamellae preparation on biological samples.
Rigort, A. et al. Focused ion beam micromachining of eukaryotic cells for cryoelectron tomography. Proc. Natl Acad. Sci. USA 109, 4449–4454 (2012).
Schaffer, M. et al. Optimized cryo-focused ion beam sample preparation aimed at in situ structural studies of membrane proteins. J. Struct. Biol. 197, 73–82 (2017).
Wagner, F. R. et al. Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat. Protoc. 15, 2041–2070 (2020).
Marko, M. et al. Focused ion beam (FIB) preparation methods for 3-D biological cryo-TEM. Microsc. Microanal. 12, 98–99 (2006).
Basgall, E. & Nicastro, D. Cryo-focused ion beam preparation of biological materials for retrieval and examination by cryo-TEM. Microsc. Microanal. 12, 1132–1133 (2006).
Hayles, M. F., Stokes, D. J., Phifer, D. & Findlay, K. C. A technique for improved focused ion beam milling of cryo-prepared life science specimens. J. Microsc. 226, 263–269 (2007).
Marko, M., Hsieh, C., Moberlychan, W., Mannella, C. A. & Frank, J. Focused ion beam milling of vitreous water: prospects for an alternative to cryo-ultramicrotomy of frozen-hydrated biological samples. J. Microsc. 222, 42–47 (2006).
Mahamid, J. et al. Visualizing the molecular sociology at the HeLa cell nuclear periphery. Science 351, 969–972 (2016). Early biological study demonstrating subtomogram averaging on FIB lamellae.
Hagen, C. et al. Structural basis of vesicle formation at the inner nuclear membrane. Cell 163, 1692–1701 (2015).
Bykov, Y. S. et al. The structure of the COPI coat determined within the cell. eLife 6, e32493 (2017).
Dodonova, S. O. et al. A structure of the COPI coat and the role of coat proteins in membrane vesicle assembly. Science 349, 195–198 (2015).
Laughlin, T. G. et al. Architecture and self-assembly of the jumbo bacteriophage nuclear shell. Nature 608, 429–435 (2022).
Pfeffer, S. et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 8, 14516 (2017).
Li, X. et al. A mammalian system for high-resolution imaging of intact cells by cryo-electron tomography. Prog. Biophys. Mol. Biol. 160, 87–96 (2021).
Kovtun, O. et al. Structure of the membrane-assembled retromer coat determined by cryo-electron tomography. Nature 561, 561–564 (2018).
Mosalaganti, S. et al. AI-based structure prediction empowers integrative structural analysis of human nuclear pores. Science 376, eabm9506 (2022).
Mosalaganti, S. et al. In situ architecture of the algal nuclear pore complex. Nat. Commun. 9, 2361 (2018).
Allegretti, M. et al. In-cell architecture of the nuclear pore and snapshots of its turnover. Nature 586, 796–800 (2020).
Zhao, Y. et al. 3D structure and in situ arrangements of CatSper channel in the sperm flagellum. Nat. Commun. 13, 3439 (2022).
Klena, N. et al. Architecture of the centriole cartwheel‐containing region revealed by cryo‐electron tomography. EMBO J. 39, e106246 (2020).
Klein, S. et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat. Commun. 11, 5885 (2020).
Berger, C. et al. Structure of the Yersinia injectisome in intracellular host cell phagosomes revealed by cryo FIB electron tomography. J. Struct. Biol. 213, 107701 (2021).
Berger, C. et al. Plasma FIB milling for the determination of structures in situ. Nat. Commun. 14, 629 (2023).
Sutton, G. et al. Assembly intermediates of orthoreovirus captured in the cell. Nat. Commun. 11, 4445 (2020).
Wang, Z. et al. The molecular basis for sarcomere organization in vertebrate skeletal muscle. Cell 184, 2135–2150.e13 (2021).
Wang, Z. et al. Structures from intact myofibrils reveal mechanism of thin filament regulation through nebulin. Science 375, 1612–1627 (2022). High-resolution in situ structures obtained on FIB lamellae, showing structural differences with structures obtained on isolated proteins.
Weiss, G. L., Kieninger, A.-K., Maldener, I., Forchhammer, K. & Pilhofer, M. Structure and function of a bacterial gap junction analog. Cell 178, 374–384.e15 (2019).
Wolff, G. et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science 369, 1395–1398 (2020). Timely study on the structural characterisation of a molecular pore formed by coronaviruses, which were not previously structurally characterized.
Zhu, H. et al. In situ structure of intestinal apical surface reveals nanobristles on microvilli. Proc. Natl Acad. Sci. USA 119, e2122249119 (2022).
Klein, S. et al. Post-correlation on-lamella cryo-CLEM reveals the membrane architecture of lamellar bodies. Commun. Biol. 4, 137 (2021).
Nickell, S., Kofler, C., Leis, A. P. & Baumeister, W. A visual approach to proteomics. Nat. Rev. Mol. Cell Biol. 7, 225–230 (2006).
Albert, S. et al. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl Acad. Sci. USA 114, 13726–13731 (2017).
Albert, S. et al. Direct visualization of degradation microcompartments at the ER membrane. Proc. Natl Acad. Sci. USA 117, 1069–1080 (2020).
Guo, Q. et al. In situ structure of neuronal C9ORF72 poly-GA aggregates reveals proteasome recruitment. Cell 172, 696–705 (2018).
Hoffman, D. P. et al. Correlative three-dimensional super-resolution and block-face electron microscopy of whole vitreously frozen cells. Science 367, eaaz5357 (2020).
Collado, J. et al. Tricalbin-mediated contact sites control ER curvature to maintain plasma membrane integrity. Dev. Cell 51, 476–487 (2019).
Khanna, K. et al. The molecular architecture of engulfment during Bacillus subtilis sporulation. eLife 8, e45257 (2019).
Böck, D. et al. In situ architecture, function, and evolution of a contractile injection system. Science 717, 713–717 (2017).
Bäuerlein, F. J. B. et al. In situ architecture and cellular interactions of PolyQ inclusions. Cell 171, 179–187.e10 (2017).
Gruber, A. et al. Molecular and structural architecture of polyQ aggregates in yeast. Proc. Natl Acad. Sci. USA 115, 201717978 (2018).
Chaikeeratisak, V. et al. Assembly of a nucleus-like structure during viral replication in bacteria. Science 355, 194–197 (2017).
Chaikeeratisak, V. et al. Viral capsid trafficking along treadmilling tubulin filaments in bacteria. Cell 177, 1771–1780.e12 (2019).
Toro-Nahuelpan, M. et al. Tailoring cryo-electron microscopy grids by photo-micropatterning for in-cell structural studies. Nat. Methods 17, 50–54 (2020).
Adrian, M., Dubochet, J., Lepault, J. & McDowall, A. W. Cryo-electron microscopy of viruses. Nature 308, 32–36 (1984).
Frederik, P. M. & Hubert, D. H. W. Cryoelectron microscopy of liposomes. Methods Enzymol. 391, 431–448 (2005).
Iancu, C. V. et al. Electron cryotomography sample preparation using the Vitrobot. Nat. Protoc. 1, 2813–2819 (2006).
Bäuerlein, F. J. B., Pastor-Pareja, J. C. & Fernández-Busnadiego, R. Cryo-electron tomography of native Drosophila tissues vitrified by plunge freezing. Preprint at bioRxiv https://doi.org/10.1101/2021.04.14.437159 (2021).
Burstein, N. L., & Maurice, D. M. Cryofixation of tissue surfaces by a propane jet for electron microscopy. Micron 9, 191–198 (1978).
Ravelli, R. B. G. et al. Cryo-EM structures from sub-nl volumes using pin-printing and jet vitrification. Nat. Commun. 11, 2563 (2020).
Escaig, J. New instruments which facilitate rapid freezing at 83K and 6K. J. Microsc. 126, 221–229 (1982).
Porta, D. & López-Iglesias, C. A comparison of cryo- versus chemical fixation in the soil green algae Jaagiella. Tissue Cell 30, 368–376 (1998).
Moor, H., Bellin, G., Sandri, C. & Akert, K. The influence of high pressure freezing on mammalian nerve tissue. Cell Tissue Res. 209, 201–216 (1980).
Harapin, J. et al. Structural analysis of multicellular organisms with cryo-electron tomography. Nat. Methods 12, 634–636 (2015).
Kelley, K. et al. Waffle method: a general and flexible approach for improving throughput in FIB-milling. Nat. Commun. 13, 1857 (2022).
Zhang, J. et al. VHUT-cryo-FIB, a method to fabricate frozen hydrated lamellae from tissue specimens for in situ cryo-electron tomography. J. Struct. Biol. 213, 107763 (2021).
Shorubalko, I., Choi, K., Stiefel, M. & Park, H. G. Ion beam profiling from the interaction with a freestanding 2D layer. Beilstein J. Nanotechnol. 8, 682–687 (2017).
Wolff, G. et al. Mind the gap: micro-expansion joints drastically decrease the bending of FIB-milled cryo-lamellae. J. Struct. Biol. 208, 107389 (2019).
Medeiros, J. M. et al. Robust workflow and instrumentation for cryo-focused ion beam milling of samples for electron cryotomography. Ultramicroscopy 190, 1–11 (2018).
Klumpe, S. et al. A modular platform for automated cryo-FIB workflows. eLife 10, e70506 (2021).
Gorelick, S., Dierickx, D., Buckley, G., Whisstock, J. & Marco, A. Assembly and imaging set up of PIE-Scope. Bio Protoc. 10, e3768 (2020).
Tacke, S. et al. A streamlined workflow for automated cryo focused ion beam milling. J. Struct. Biol. 213, 107743 (2021). An implementation of automated lamella fabrication, and important improvements on the FIB/SEM microscope to minimize ice growth on lamellae.
Rubino, S. et al. A site-specific focused-ion-beam lift-out method for cryo transmission electron microscopy. J. Struct. Biol. 180, 572–576 (2012).
Mahamid, J. et al. A focused ion beam milling and lift-out approach for site-specific preparation of frozen-hydrated lamellas from multicellular organisms. J. Struct. Biol. 192, 262–269 (2015).
Parmenter, C. D. & Nizamudeen, Z. A. Cryo-FIB-lift-out: practically impossible to practical reality. J. Microsc. 281, 157–174 (2021).
De Winter, D. A. M., Hsieh, C., Marko, M. & Hayles, M. F. Cryo-FIB preparation of whole cells and tissue for cryo-TEM: use of high-pressure frozen specimens in tubes and planchets. J. Microsc. 281, 125–137 (2021).
Schaffer, M. et al. A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat. Methods 16, 757–762 (2019). Demonstration of the lift-out technique on small organisms using a cryo-gripper, resulting in lamellae with sufficient quality for subtomogram averaging of ribosomes.
Fuest, M. et al. In situ microfluidic cryofixation for cryo focused ion beam milling and cryo electron tomography. Sci. Rep. 9, 19133 (2019).
Rigort, A. et al. Micromachining tools and correlative approaches for cellular cryo-electron tomography. J. Struct. Biol. 172, 169–179 (2010).
Gorelick, S. et al. PIE-scope, integrated cryo-correlative light and FIB/SEM microscopy. eLife 8, e45919 (2019). Integration of a fluorescence microscope in a cryo-FIB/SEM microscope for CLEM.
Burnett, T. L. et al. Large volume serial section tomography by Xe plasma FIB dual beam microscopy. Ultramicroscopy 161, 119–129 (2016).
Liu, J. et al. Effect of ion irradiation introduced by focused ion-beam milling on the mechanical behaviour of sub-micron-sized samples. Sci. Rep. 10, 10324 (2020).
Grigorieff, N. Direct detection pays off for electron cryo-microscopy. eLife 2, 2–4 (2013).
Danev, R., Buijsse, B., Khoshouei, M., Plitzko, J. M. & Baumeister, W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc. Natl Acad. Sci. USA 111, 15635–15640 (2014).
Turoňová, B. et al. Benchmarking tomographic acquisition schemes for high-resolution structural biology. Nat. Commun. 11, 876 (2020).
Schwartz, O. et al. Laser phase plate for transmission electron microscopy. Nat. Methods 16, 1016–1020 (2019).
Hagen, W. J. H., Wan, W. & Briggs, J. A. G. Implementation of a cryo-electron tomography tilt-scheme optimized for high resolution subtomogram averaging. J. Struct. Biol. 197, 191–198 (2017). Implementation of a dose-symmetric tilt scheme, which has rapidly become the standard for cryo-electron tomography.
Chreifi, G., Chen, S., Metskas, L. A., Kaplan, M. & Jensen, G. J. Rapid tilt-series acquisition for electron cryotomography. J. Struct. Biol. 205, 163–169 (2019).
Eisenstein, F., Danev, R. & Pilhofer, M. Improved applicability and robustness of fast cryo-electron tomography data acquisition. J. Struct. Biol. 208, 107–114 (2019).
Eisenstein, F. et al. Parallel cryo electron tomography on in situ lamellae. Nat. Methods 20, 131–138 (2023).
Yang, J. E. et al. Correlative cryogenic montage electron tomography for comprehensive in-situ whole-cell structural studies. Preprint at bioRxiv https://doi.org/10.1101/2021.12.31.474669 (2022).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Bharat, T. A. M. & Scheres, S. H. W. Resolving macromolecular structures from electron cryo-tomography data using subtomogram averaging in RELION. Nat. Protoc. 11, 2054–2065 (2016). Reliable software for cryo-EM data processing, which has increasingly become more suitable for subtomogram averaging.
Fernández, J. J., Li, S. & Crowther, R. A. CTF determination and correction in electron cryotomography. Ultramicroscopy 106, 587–596 (2006).
Mastronarde, D. N. & Held, S. R. Automated tilt series alignment and tomographic reconstruction in IMOD. J. Struct. Biol. 197, 102–113 (2017). IMOD has been an invaluable tool for tomogram reconstructions and visualization for many years.
Turoňová, B., Schur, F. K. M., Wan, W. & Briggs, J. A. G. Efficient 3D-CTF correction for cryo-electron tomography using NovaCTF improves subtomogram averaging resolution to 3.4Å. J. Struct. Biol. 199, 187–195 (2017).
Kunz, M. & Frangakis, A. S. Three-dimensional CTF correction improves the resolution of electron tomograms. J. Struct. Biol. 197, 114–122 (2017).
Himes, B. A. & Zhang, P. emClarity: software for high-resolution cryo-electron tomography and subtomogram averaging. Nat. Methods 15, 955–961 (2018).
Chen, M. et al. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods 16, 1161–1168 (2019).
Tegunov, D., Xue, L., Dienemann, C., Cramer, P. & Mahamid, J. Multi-particle cryo-EM refinement with M visualizes ribosome-antibiotic complex at 3.5Å in cells. Nat. Methods 18, 186–193 (2021). Subtomogram averaging with Warp, RELION and M allowed for pseudo-atomic structures of ribosomes to be obtained in vivo.
Berger, C., Ravelli, R. B. G., López-Iglesias, C. & Peters, P. J. Endocytosed nanogold fiducials for improved in-situ cryo-electron tomography tilt-series alignment. J. Struct. Biol. 213, 107698 (2021).
Castaño-Díez, D. The Dynamo package for tomography and subtomogram averaging: components for MATLAB, GPU computing and EC2 Amazon Web Services. Acta Crystallogr. D Struct. Biol. 73, 478–487 (2017).
Song, K. et al. In situ structure determination at nanometer resolution using TYGRESS. Nat. Methods 17, 201–208 (2020).
Sanchez, R. M., Zhang, Y., Chen, W., Dietrich, L. & Kudryashev, M. Subnanometer-resolution structure determination in situ by hybrid subtomogram averaging—single particle cryo-EM. Nat. Commun. 11, 3709 (2020).
Song, K. et al. In situ localization of N and C termini of subunits of the flagellar nexin–dynein regulatory complex (N–DRC) using SNAP tag and cryo-electron tomography. J. Biol. Chem. 290, 5341–5353 (2015).
Yi, H. et al. Native immunogold labeling of cell surface proteins and viral glycoproteins for cryo-electron microscopy and cryo-electron tomography applications. J. Histochem. Cytochem. 63, 780–792 (2015).
Spehner, D. et al. Cryo-FIB–SEM as a promising tool for localizing proteins in 3D. J. Struct. Biol. 211, 107528 (2020).
Silvester, E. et al. DNA origami signposts for identifying proteins on cell membranes by electron cryotomography. Cell 184, 1110–1121.e16 (2021).
Wang, Q., Mercogliano, C. P. & Löwe, J. A ferritin-based label for cellular electron cryotomography. Structure 19, 147–154 (2011).
Frangakis, A. S. It’s noisy out there! A review of denoising techniques in cryo-electron tomography. J. Struct. Biol. 213, 107804 (2021).
Frangakis, A. S. & Hegerl, R. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J. Struct. Biol. 135, 239–250 (2001).
Croxford, M. et al. Entropy-regularized deconvolution of cellular cryotransmission electron tomograms. Proc. Natl Acad. Sci. USA 118, e2108738118 (2021).
Bepler, T., Kelley, K., Noble, A. J. & Berger, B. Topaz-denoise: general deep denoising models for cryoEM and cryoET. Nat. Commun. 11, 5208 (2020).
Buchholz, T.-O. et al. Content-aware image restoration for electron microscopy. Methods Cell Biol. 152, 277–289 (2019).
Tegunov, D. & Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 16, 1146–1152 (2019).
Fernandez, J.-J. et al. Removing contamination-induced reconstruction artifacts from cryo-electron tomograms. Biophys. J. 110, 850–859 (2016).
Wagner, T. et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019).
Chen, M. et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods 14, 983–985 (2017).
Moebel, E. et al. Deep learning improves macromolecule identification in 3D cellular cryo-electron tomograms. Nat. Methods 18, 1386–1394 (2021).
Xu, M. et al. De novo structural pattern mining in cellular electron cryotomograms. Structure 27, 679–691.e14 (2019).
Martinez-Sanchez, A. et al. Template-free detection and classification of membrane-bound complexes in cryo-electron tomograms. Nat. Methods 17, 209–216 (2020).
Martinez-Sanchez, A., Garcia, I., Asano, S., Lucic, V. & Fernandez, J. J. Robust membrane detection based on tensor voting for electron tomography. J. Struct. Biol. 186, 49–61 (2014).
Salfer, M., Collado, J. F., Baumeister, W., Fernández-Busnadiego, R. & Martínez-Sánchez, A. Reliable estimation of membrane curvature for cryo-electron tomography. PLoS Comput. Biol. 16, e1007962 (2020).
Frangakis, A. S. Mean curvature motion facilitates the segmentation and surface visualization of electron tomograms. J. Struct. Biol. 214, 107833 (2022).
Deerinck, T. J. et al. Fluorescence photooxidation with eosin: a method for high resolution immunolocalization and in situ hybridization detection for light and electron microscopy. J. Cell Biol. 126, 901–910 (1994).
Schwartz, C. L., Sarbash, V. I., Atallakhanov, F. I., Mcintosh, J. R. & Nicastro, D. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J. Microsc. 227, 98–109 (2007).
Sartori, A. et al. Correlative microscopy: bridging the gap between fluorescence light microscopy and cryo-electron tomography. J. Struct. Biol. 160, 135–145 (2007).
Jasnin, M. et al. The architecture of traveling actin waves revealed by cryo-electron tomography. Structure 27, 1211–1223.e5 (2019).
Chakraborty, S., Mahamid, J. & Baumeister, W. Cryoelectron tomography reveals nanoscale organization of the cytoskeleton and its relation to microtubule curvature inside cells. Structure 28, 991–1003.e4 (2020).
Koning, R. I. et al. MAVIS: an integrated system for live microscopy and vitrification. Ultramicroscopy 143, 67–76 (2014).
Van Driel, L. F., Valentijn, J. A., Valentijn, K. M., Koning, R. I. & Koster, A. J. Tools for correlative cryo-fluorescence microscopy and cryo-electron tomography applied to whole mitochondria in human endothelial cells. Eur. J. Cell Biol. 88, 669–684 (2009).
Schorb, M. & Briggs, J. A. G. Correlated cryo-fluorescence and cryo-electron microscopy with high spatial precision and improved sensitivity. Ultramicroscopy 143, 24–32 (2014).
Arnold, J. et al. Site-specific cryo-focused ion beam sample preparation guided by 3D correlative microscopy. Biophys. J. 110, 860–869 (2016).
Li, S. et al. ELI trifocal microscope: a precise system to prepare target cryo-lamellae for in situ cryo-ET study. Nat. Methods https://doi.org/10.1038/s41592-022-01748-0 (2023).
Derosier, D. J. Where in the cell is my protein?. Q. Rev. Biophys. 54, e9 (2021).
Boltje, D. B. et al. A cryogenic, coincident fluorescence, electron and ion beam microscope. eLife 11, e82891 (2022).
Wu, G.-H. et al. Multi-scale 3D cryo-correlative microscopy for vitrified cells. Structure 28, 1231–1237.e3 (2020).
Nahmani, M., Lanahan, C., DeRosier, D. & Turrigiano, G. G. High-numerical-aperture cryogenic light microscopy for increased precision of superresolution reconstructions. Proc. Natl Acad. Sci. USA 114, 3832–3836 (2017).
Faoro, R. et al. Aberration-corrected cryoimmersion light microscopy. Proc. Natl Acad. Sci. USA 115, 1204–1209 (2018).
Wang, L. et al. Solid immersion microscopy images cells under cryogenic conditions with 12 nm resolution. Commun. Biol. 2, 74 (2019).
Betzig, E. et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 (2006).
Klar, T. A., Jakobs, S., Dyba, M., Egner, A. & Hell, S. W. Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl Acad. Sci. USA 97, 8206–8210 (2000).
Dickson, R. M., Cubitt, A. B., Tsien, R. Y. & Moerner, W. E. On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 388, 355–358 (1997).
Kaufmann, R. et al. Super-resolution microscopy using standard fluorescent proteins in intact cells under cryo-conditions. Nano Lett. 14, 4171–4175 (2014).
Chang, Y.-W. et al. Correlated cryogenic photoactivated localization microscopy and cryo-electron tomography. Nat. Methods 11, 737–739 (2014).
Liu, B. et al. Three-dimensional super-resolution protein localization correlated with vitrified cellular context. Sci. Rep. 5, 13017 (2015).
Dahlberg, P. D. et al. Cryogenic single-molecule fluorescence annotations for electron tomography reveal in situ organization of key proteins in Caulobacter. Proc. Natl Acad. Sci. USA 117, 202001849 (2020).
Tuijtel, M. W., Koster, A. J., Jakobs, S., Faas, F. G. A. & Sharp, T. H. Correlative cryo super-resolution light and electron microscopy on mammalian cells using fluorescent proteins. Sci. Rep. 9, 1369 (2019). Study demonstrating localization-based super-resolution cryo-fluorescence microscopy on mammalian cells and characterization of the light intensities that can be used without devitrifying samples.
Moser, F. et al. Cryo-SOFI enabling low-dose super-resolution correlative light and electron cryo-microscopy. Proc. Natl Acad. Sci. USA 116, 4804–4809 (2019).
Phillips, M. A. et al. CryoSIM: super-resolution 3D structured illumination cryogenic fluorescence microscopy for correlated ultrastructural imaging. Optica 7, 802–812 (2020).
Sexton, D. L., Burgold, S., Schertel, A. & Tocheva, E. I. Super-resolution confocal cryo-CLEM with cryo-FIB milling for in situ imaging of Deinococcus radiodurans. Curr. Res. Struct. Biol. 4, 1–9 (2022).
Gupta, H., McCann, M. T., Donati, L. & Unser, M. CryoGAN: a new reconstruction paradigm for single-particle cryo-EM via deep adversarial learning. IEEE Trans. Comput. Imaging 7, 759–774 (2021).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Paul-Gilloteaux, P. et al. eC-CLEM: flexible multidimensional registration software for correlative microscopies. Nat. Methods 14, 102–103 (2017).
Acknowledgements
We thank the Microscopy CORE Lab of the Maastricht MultiModal Molecular Imaging Institute of the Faculty of Health, Medicine and Life Sciences at Maastricht University for technical and scientific support. We thank M. Bárcena and S. Raunser and their co-authors for permission to modify and republish their figures. We thank H. Nguyen for editing the manuscript. We thank A. McDowall for critical reading of the manuscript. We acknowledge co-funding by the PPP Allowance made available by Health ~ Holland, Top Sector Life Sciences & Health, to stimulate public–private partnerships, under project number LHSM18067, as well as from the Netherlands Organisation for Scientific Research (NWO) in the framework of National Roadmap NEMI project number 184.034.014.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The University of Maastricht has filed patents, with R.B.G.R., C.L.-I. and P.J.P. as inventors, regarding vitrification devices. P.J.P. is a shareholder and chief scientific officer of the start-up CryoSol-World, which holds the licenses of these submitted patents (WO 2017/220750 and EP 3260839 B1). The other authors declare no competing interests.
Peer review
Peer review information
Nature Methods thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Berger, C., Premaraj, N., Ravelli, R.B.G. et al. Cryo-electron tomography on focused ion beam lamellae transforms structural cell biology. Nat Methods 20, 499–511 (2023). https://doi.org/10.1038/s41592-023-01783-5
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-023-01783-5
This article is cited by
-
Dynamins maintain nuclear envelope homeostasis and genome stability
Nature Communications (2026)
-
Correlative cryogenic light and electron tomography of eukaryotic cells
Nature Reviews Methods Primers (2025)
-
Integrating diverse experimental information to assist protein complex structure prediction by GRASP
Nature Methods (2025)
-
Rapid structural analysis of bacterial ribosomes in situ
Communications Biology (2025)
-
Design and Performance Analysis of a Multilayer Photonic Crystal Biosensor for Enhanced Detection of Melanoma Skin Cancer
Sensing and Imaging (2025)