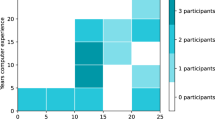
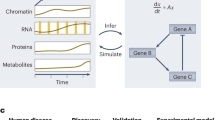
Abstract
Major computational challenges exist in relation to the collection, curation, processing and analysis of large genomic and imaging datasets, as well as the simulation of larger and more realistic models in systems biology. Here we discuss how a relative newcomer among programming languages—Julia—is poised to meet the current and emerging demands in the computational biosciences and beyond. Speed, flexibility, a thriving package ecosystem and readability are major factors that make high-performance computing and data analysis available to an unprecedented degree. We highlight how Julia’s design is already enabling new ways of analyzing biological data and systems, and we provide a list of resources that can facilitate the transition into Julian computing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Change history
29 April 2023
A Correction to this paper has been published: https://doi.org/10.1038/s41592-023-01887-y
References
Tomlin, C. J. & Axelrod, J. D. Biology by numbers: mathematical modelling in developmental biology. Nat. Rev. Genet. 8, 331–340 (2007).
Auton, A. et al. A global reference for human genetic variation. Nature 526, 68–74 (2015).
Robson, B. Computers and viral diseases. preliminary bioinformatics studies on the design of a synthetic vaccine and a preventative peptidomimetic antagonist against the SARS-CoV-2 (2019-nCoV, COVID-19) coronavirus. Comput. Biol. Med. 119, 103670 (2020).
Seefeld, K. & Linder, E. Statistics Using R with Biological Examples (K. Seefeld, 2007).
Ekmekci, B., McAnany, C. E. & Mura, C. An introduction to programming for bioscientists: a Python-based primer. PLoS Comput. Biol. 12, e1004867 (2016).
Sengupta, A. & Edelman, A. Julia High Performance (Packt Publishing, 2019).
Nazarathy, Y. & Klok, H. Statistics with Julia: Fundamentals for Data Science, Machine Learning and Artificial Intelligence (Springer, 2021).
Bezanson, J., Edelman, A., Karpinski, S. & Shah, V. B. Julia: a fresh approach to numerical computing. SIAM Rev. 59, 65–98 (2017).
Lauwens, B. & Downey, A. Think Julia: How to Think like a Computer Scientist (O’Reilly Media, 2021).
Marx, V. The big challenges of big data. Nature 498, 255–260 (2013).
Björnsson, B. et al. Digital twins to personalize medicine. Genome Med. 12, 4 (2019).
Laubenbacher, R., Sluka, J. P. & Glazier, J. A. Using digital twins in viral infection. Science 371, 1105–1106 (2021).
Chan, T. E., Stumpf, M. P. & Babtie, A. C. Gene regulatory network inference from single-cell data using multivariate information measures. Cell Syst. 5, 251–267.e3 (2017).
Tankhilevich, E. et al. GpABC: a Julia package for approximate Bayesian computation with Gaussian process emulation. Bioinformatics 36, 3286–3287 (2020).
Innes, M. Flux: elegant machine learning with Julia. J. Open Source Softw. 3, 602 (2018).
Rackauckas, C. & Nie, Q. DifferentialEquations.jl—a performant and feature-rich ecosystem for solving differential equations in Julia. J. Open Res. Softw. 5, 15 (2017).
Chen, J. et al. Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat. Protoc. 12, 566–580 (2017).
Mahon, S. S. M. et al. Information theory and signal transduction systems: from molecular information processing to network inference. Semin. Cell Dev. Biol. 35, 98–108 (2014).
Meyer, P. E., Lafitte, F. & Bontempi, G. minet: a R/Bioconductor package for inferring large transcriptional networks using mutual information. BMC Bioinformatics 9, 461 (2008).
Bates, D. Julia MixedModels from R. https://rpubs.com/dmbates/377897 (2018).
Lange, K. Algorithms from the Book (SIAM, 2020).
Oliveira, S. & Stewart, D. E. Writing Scientific Software: a Guide to Good Style (Cambridge Univ. Press, 2006).
Alyass, A., Turcotte, M. & Meyre, D. From big data analysis to personalized medicine for all: challenges and opportunities. BMC Med. Genom. 8, 33 (2015).
Gomez-Cabrero, D. et al. Data integration in the era of omics: current and future challenges. BMC Syst. Biol. 8, I1 (2014).
Greener, J. G., Selvaraj, J. & Ward, B. J. BioStructures.jl: read, write and manipulate macromolecular structures in julia. Bioinformatics 36, 4206–4207 (2020).
Rego, N. & Koes, D. 3Dmol.js: molecular visualization with WebGL. Bioinformatics 31, 1322–1324 (2014).
Hayashi, T. et al. Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs. Nat. Commun. 9, 619 (2018).
Greener, J. G., Filippis, I. & Sternberg, M. J. Predicting protein dynamics and allostery using multi-protein atomic distance constraints. Structure 25, 546–558 (2017).
Zea, D. J., Anfossi, D., Nielsen, M. & Marino-Buslje, C. MIToS.jl: mutual information tools for protein sequence analysis in the Julia language. Bioinformatics 33, 564–565 (2017).
Cock, P. J. A. et al. Biopython: freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009).
Kunzmann, P. & Hamacher, K. Biotite: a unifying open source computational biology framework in Python. BMC Bioinformatics 19, 346 (2018).
Perera, R. Programming languages for interactive computing. Electron. Notes Theor. Comput. Sci. 203, 35–52 (2008).
Kirk, P. D. W., Babtie, A. C. & Stumpf, M. P. H. Systems biology (un)certainties. Science 350, 386–388 (2015).
Kirk, P., Thorne, T. & Stumpf, M. P. Model selection in systems and synthetic biology. Curr. Opin. Biotechnol. 24, 767–774 (2013).
Warne, D. J., Baker, R. E. & Simpson, M. J. Simulation and inference algorithms for stochastic biochemical reaction networks: from basic concepts to state-of-the-art. J. R. Soc. Interface 16, 20180943 (2019).
Filippi, S. et al. Robustness of MEK-ERK dynamics and origins of cell-to-cell variability in MAPK signaling. Cell Rep. 15, 2524–2535 (2016).
Michailovici, I. et al. Nuclear to cytoplasmic shuttling of ERK promotes differentiation of muscle stem/progenitor cells. Development 141, 2611–2620 (2014).
MacLean, A. L., Rosen, Z., Byrne, H. M. & Harrington, H. A. Parameter-free methods distinguish Wnt pathway models and guide design of experiments. Proc. Natl Acad. Sci. USA 112, 2652–2657 (2015).
Loman, T. E. et al. Catalyst: fast biochemical modeling with Julia. Preprint at bioRxiv https://doi.org/10.1101/2022.07.30.502135 (2022).
Harrington, H. A., Feliu, E., Wiuf, C. & Stumpf, M. P. Cellular compartments cause multistability and allow cells to process more information. Biophys. J. 104, 1824–1831 (2013).
Mogensen, P. K. & Riseth, A. N. Optim: a mathematical optimization package for Julia. J. Open Source Softw. 3, 615 (2018).
Dunning, I., Huchette, J. & Lubin, M. JuMP: a modeling language for mathematical optimization. SIAM Rev. 59, 295–320 (2017).
Ge, H., Xu, K. & Ghahramani, Z. Turing: a language for flexible probabilistic inference. In Proc. 21st International Conference on Artificial Intelligence and Statistics 1682–1690 (Proc. Machine Learning Res., 2018).
Liepe, J. et al. A framework for parameter estimation and model selection from experimental data in systems biology using approximate bayesian computation. Nat. Protoc. 9, 439–456 (2014).
Harris, C. R. et al. Array programming with NumPy. Nature 585, 357–362 (2020).
Stanitzki, M. & Strube, J. Performance of Julia for high energy physics analyses. Comput. Softw. Big Sci. 5, 10 (2021).
Rackauckas, C. et al. Accelerated predictive healthcare analytics with Pumas, a high performance pharmaceutical modeling and simulation platform. Preprint at bioRxiv https://doi.org/10.1101/2020.11.28.402297 (2020).
Whitney, T. & Taylor, V. Increasing women and underrepresented minorities in computing: the landscape and what you can do. Computer 51, 24–31 (2018).
Sharpe, J. Computer modeling in developmental biology: growing today, essential tomorrow. Development 144, 4214–4225 (2017).
Rackauckas, C. Benchmark of ODE solvers in Julia. https://github.com/SciML/MATLABDiffEq.jl (2019).
Acknowledgements
We thank all attendees of the Birds of a Feather session Julia for Biologists at JuliaCon2021; D. F. Gleich for allowing us to run an experiment on his servers; and R. Patro for discussions about Rust. E.R. acknowledges financial support through a University of Melbourne PhD scholarship. A.L.M. acknowledges support from the National Science Foundation (DMS 2045327). T.E.H. acknowledges NIH 1UF1NS108176. The information, data and work presented herein was funded in part by the Advanced Research Projects Agency—Energy under award numbers DE-AR0001222 and DE-AR0001211, as well as National Science Foundation award number IIP-1938400. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the US Government or any agency thereof. M.P.H.S. acknowledges funding from the University of Melbourne Driving Research Momentum initiative and Volkswagen Foundation Life? program grant (grant number 93063), as well as support through an Australian Research Council Laureate Fellowship.
Author information
Authors and Affiliations
Contributions
E.R. and M.P.H.S. conceived of the concept of the project and were in charge of the overall direction and planning. All authors contributed to writing the manuscript and have read and approved the final version.
Corresponding author
Ethics declarations
Competing interests
E.R. is a Sales Engineer at JuliaHub. C.R. is the Vice President of Modeling and Simulation at JuliaHub and Director of Scientific Research at Pumas-AI. T.E.H. is a steward of the Julia project. H.N. is a Senior Computer Scientist at RelationalAI. J.G.G., A.L.M. and M.P.H.S. declare no competing interests.
Peer review
Peer review information
Nature Methods thanks Nico Stuurman and the other, anonymous, reviewers for their contribution to the peer review of this work. Primary Handling Editor: Rita Strack, in collaboration with the Nature Methods team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Examples and Supplementary Table 1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Roesch, E., Greener, J.G., MacLean, A.L. et al. Julia for biologists. Nat Methods 20, 655–664 (2023). https://doi.org/10.1038/s41592-023-01832-z
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41592-023-01832-z
This article is cited by
-
Translating microbial kinetics into quantitative responses and testable hypotheses using Kinbiont
Nature Communications (2025)
-
RankCompV3: a differential expression analysis algorithm based on relative expression orderings and applications in single-cell RNA transcriptomics
BMC Bioinformatics (2024)
-
Developmental hematopoietic stem cell variation explains clonal hematopoiesis later in life
Nature Communications (2024)